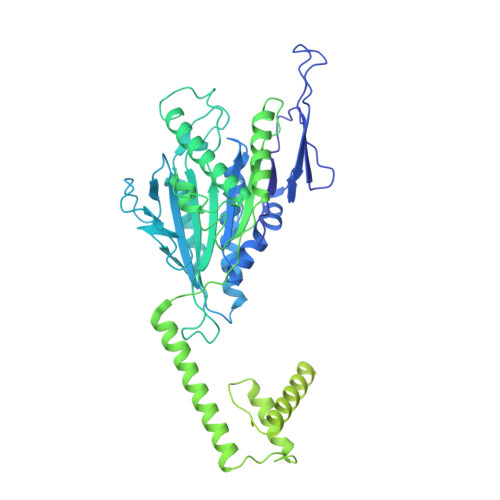
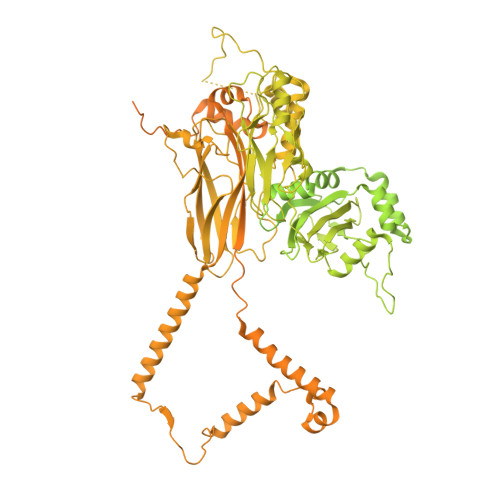
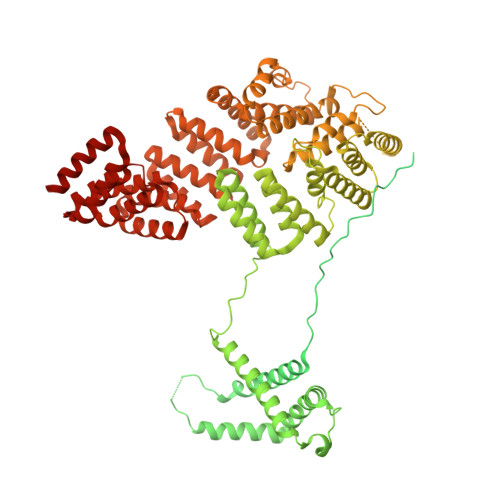
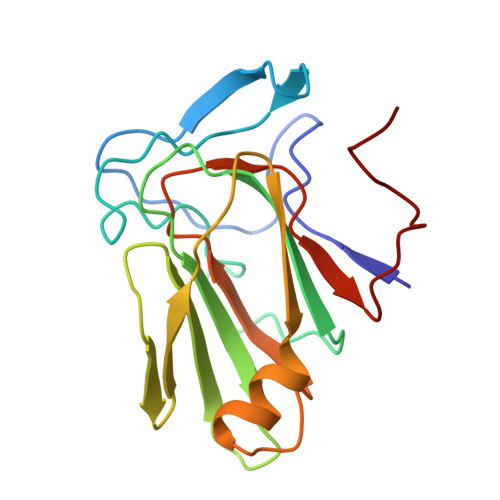
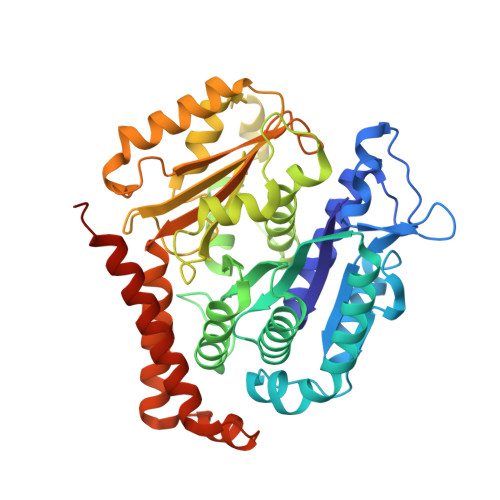
Cryo-EM structure of an active central apparatus.
Han, L., Rao, Q., Yang, R., Wang, Y., Chai, P., Xiong, Y., Zhang, K.(2022) Nat Struct Mol Biol 29: 472-482
- PubMed: 35578022
- DOI: https://doi.org/10.1038/s41594-022-00769-9
- Primary Citation of Related Structures:
7N61, 7N6G - PubMed Abstract:
Accurately regulated ciliary beating in time and space is critical for diverse cellular activities, which impact the survival and development of nearly all eukaryotic species. An essential beating regulator is the conserved central apparatus (CA) of motile cilia, composed of a pair of microtubules (C1 and C2) associated with hundreds of protein subunits per repeating unit. It is largely unclear how the CA plays its regulatory roles in ciliary motility. Here, we present high-resolution structures of Chlamydomonas reinhardtii CA by cryo-electron microscopy (cryo-EM) and its dynamic conformational behavior at multiple scales. The structures show how functionally related projection proteins of CA are clustered onto a spring-shaped scaffold of armadillo-repeat proteins, facilitated by elongated rachis-like proteins. The two halves of the CA are brought together by elastic chain-like bridge proteins to achieve coordinated activities. We captured an array of kinesin-like protein (KLP1) in two different stepping states, which are actively correlated with beating wave propagation of cilia. These findings establish a structural framework for understanding the role of the CA in cilia.
- Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, CT, USA.
Organizational Affiliation: