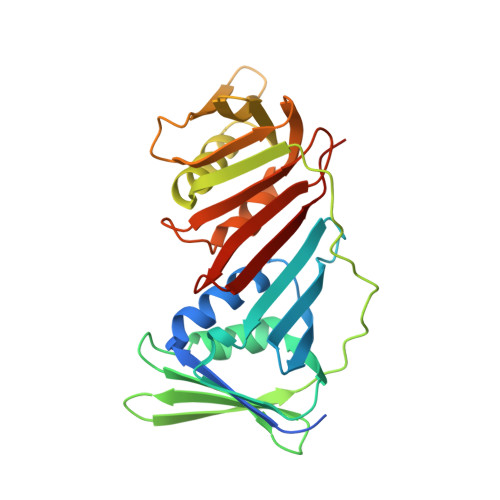
PCNA from Thermococcus gammatolerans: A protein involved in chromosomal DNA metabolism intrinsically resistant at high levels of ionizing radiation.
Marin-Tovar, Y., Serrano-Posada, H., Diaz-Vilchis, A., Rudino-Pinera, E.(2022) Proteins 90: 1684-1698
- PubMed: 35435259
- DOI: https://doi.org/10.1002/prot.26346
- Primary Citation of Related Structures:
7N5I, 7N5J, 7N5K, 7N5L, 7N5M, 7N5N - PubMed Abstract:
Proliferating cell nuclear antigen (PCNA) is an essential protein for cell viability in archaea and eukarya, since it is involved in DNA replication and repair. In order to obtain insights regarding the characteristics that confer radioresistance, the structural study of the PCNA from Thermococcus gammatolerans (PCNA Tg ) in a gradient of ionizing radiation by X-ray crystallography was carried out, together with a bioinformatic analysis of homotrimeric PCNA structures, their sequences, and their molecular interactions. The results obtained from the datasets and the accumulated radiation dose for the last collection from three crystals revealed moderate and localized damage, since even with the loss of resolution, the electron density map corresponding to the last collection allowed to build the whole structure. Attempting to understand this behavior, multiple sequence alignments, and structural superpositions were performed, revealing that PCNA is a protein with a poorly conserved sequence, but with a highly conserved structure. The PCNA Tg presented the highest percentage of charged residues, mostly negatively charged, with a proportion of glutamate more than double aspartate, lack of cysteines and tryptophan, besides a high number of salt bridges. The structural study by X-ray crystallography reveals that the PCNA Tg has the intrinsic ability to resist high levels of ionizing radiation, and the bioinformatic analysis suggests that molecular evolution selected a particular composition of amino acid residues, and their consequent network of synergistic interactions for extreme conditions, as a collateral effect, conferring radioresistance to a protein involved in the chromosomal DNA metabolism of a radioresistant microorganism.
- Laboratorio de Bioquímica Estructural, Departamento de Medicina Molecular y Bioprocesos, Instituto de Biotecnología (IBt), Universidad Nacional Autónoma de México (UNAM), Cuernavaca, Mexico.
Organizational Affiliation: