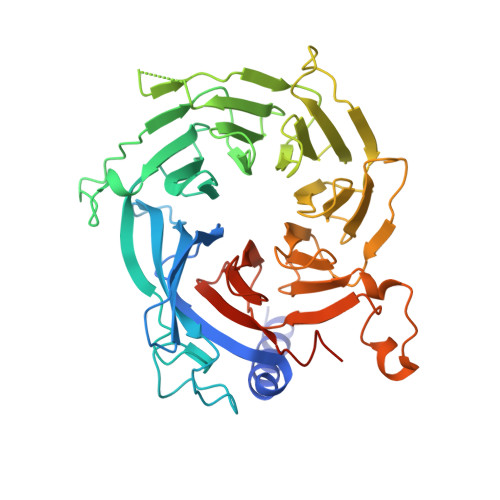
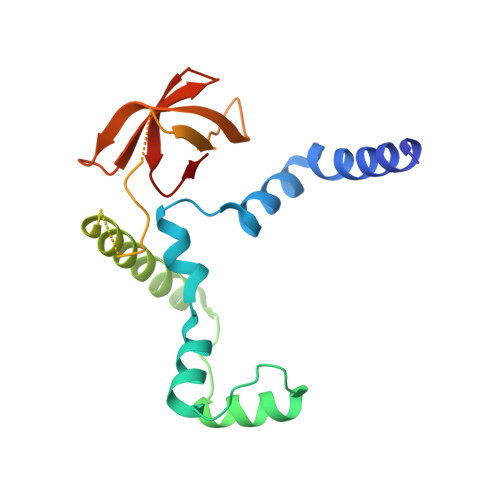
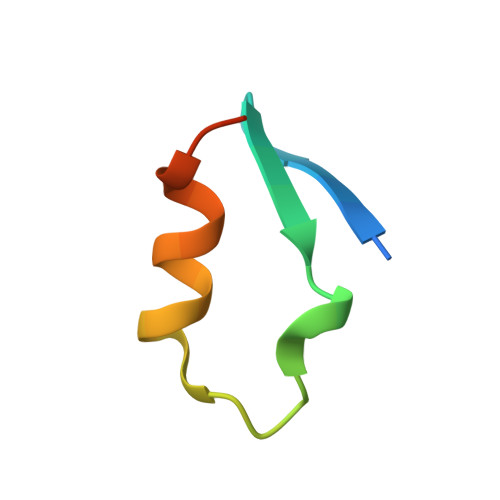
The MuvB complex binds and stabilizes nucleosomes downstream of the transcription start site of cell-cycle dependent genes.
Asthana, A., Ramanan, P., Hirschi, A., Guiley, K.Z., Wijeratne, T.U., Shelansky, R., Doody, M.J., Narasimhan, H., Boeger, H., Tripathi, S., Muller, G.A., Rubin, S.M.(2022) Nat Commun 13: 526-526
- PubMed: 35082292
- DOI: https://doi.org/10.1038/s41467-022-28094-1
- Primary Citation of Related Structures:
7N40 - PubMed Abstract:
The chromatin architecture in promoters is thought to regulate gene expression, but it remains uncertain how most transcription factors (TFs) impact nucleosome position. The MuvB TF complex regulates cell-cycle dependent gene-expression and is critical for differentiation and proliferation during development and cancer. MuvB can both positively and negatively regulate expression, but the structure of MuvB and its biochemical function are poorly understood. Here we determine the overall architecture of MuvB assembly and the crystal structure of a subcomplex critical for MuvB function in gene repression. We find that the MuvB subunits LIN9 and LIN37 function as scaffolding proteins that arrange the other subunits LIN52, LIN54 and RBAP48 for TF, DNA, and histone binding, respectively. Biochemical and structural data demonstrate that MuvB binds nucleosomes through an interface that is distinct from LIN54-DNA consensus site recognition and that MuvB increases nucleosome occupancy in a reconstituted promoter. We find in arrested cells that MuvB primarily associates with a tightly positioned +1 nucleosome near the transcription start site (TSS) of MuvB-regulated genes. These results support a model that MuvB binds and stabilizes nucleosomes just downstream of the TSS on its target promoters to repress gene expression.
- Department of Chemistry and Biochemistry, University of California, Santa Cruz, CA, 95064, USA.
Organizational Affiliation: