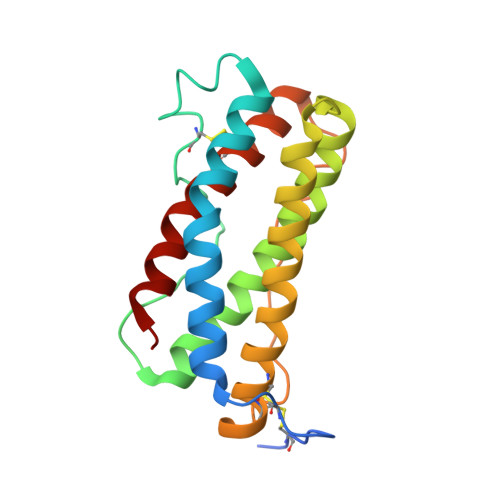
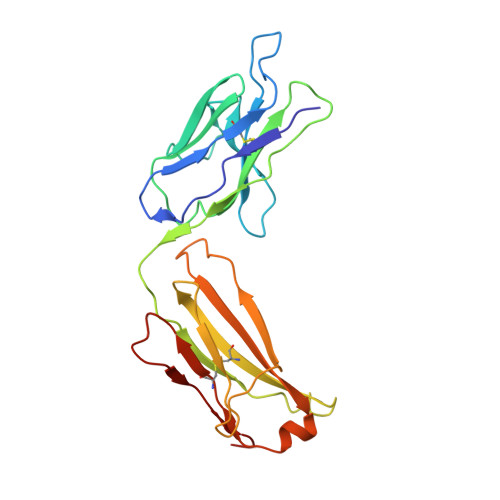
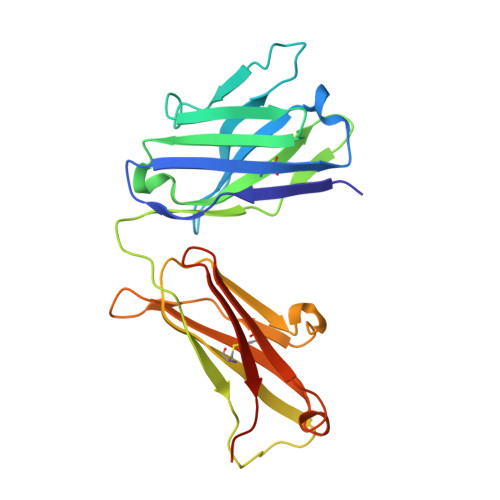
Therapeutic Targeting of LIF Overcomes Macrophage-mediated Immunosuppression of the Local Tumor Microenvironment.
Hallett, R.M., Bonfill-Teixidor, E., Iurlaro, R., Arias, A., Raman, S., Bayliss, P., Egorova, O., Neva-Alejo, A., McGray, A.R., Lau, E., Bosch, A., Beilschmidt, M., Maetzel, D., Fransson, J., Huber-Ruano, I., Anido, J., Julien, J.P., Giblin, P., Seoane, J.(2023) Clin Cancer Res 29: 791-804
- PubMed: 36441800
- DOI: https://doi.org/10.1158/1078-0432.CCR-21-1888
- Primary Citation of Related Structures:
7N0A - PubMed Abstract:
Leukemia inhibitory factor (LIF) is a multifunctional cytokine with numerous reported roles in cancer and is thought to drive tumor development and progression. Characterization of LIF and clinical-stage LIF inhibitors would increase our understanding of LIF as a therapeutic target. We first tested the association of LIF expression with transcript signatures representing multiple processes regulating tumor development and progression. Next, we developed MSC-1, a high-affinity therapeutic antibody that potently inhibits LIF signaling and tested it in immune competent animal models of cancer. LIF was associated with signatures of tumor-associated macrophages (TAM) across 7,769 tumor samples spanning 22 solid tumor indications. In human tumors, LIF receptor was highly expressed within the macrophage compartment and LIF treatment drove macrophages to acquire immunosuppressive capacity. MSC-1 potently inhibited LIF signaling by binding an epitope that overlaps with the gp130 receptor binding site on LIF. MSC-1 showed monotherapy efficacy in vivo and drove TAMs to acquire antitumor and proinflammatory function in syngeneic colon cancer mouse models. Combining MSC-1 with anti-PD1 leads to strong antitumor response and a long-term tumor-free survival in a significant proportion of treated mice. Overall, our findings highlight LIF as a therapeutic target for cancer immunotherapy.
- Northern Biologics, Toronto, Ontario, Canada.
Organizational Affiliation: