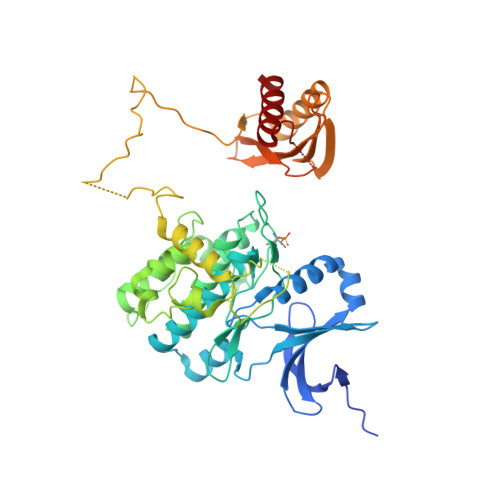
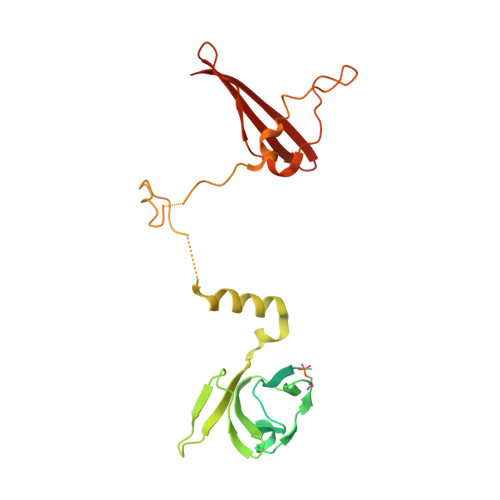
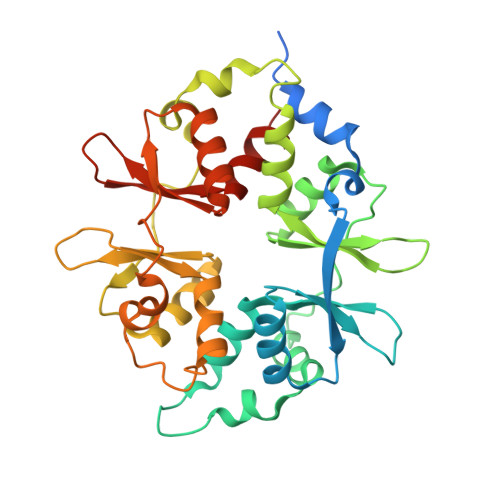
Structure-function analysis of the AMPK activator SC4 and identification of a potent pan AMPK activator.
Ovens, A.J., Gee, Y.S., Ling, N.X.Y., Yu, D., Hardee, J.P., Chung, J.D., Ngoei, K.R.W., Waters, N.J., Hoffman, N.J., Scott, J.W., Loh, K., Spengler, K., Heller, R., Parker, M.W., Lynch, G.S., Huang, F., Galic, S., Kemp, B.E., Baell, J.B., Oakhill, J.S., Langendorf, C.G.(2022) Biochem J 479: 1181-1204
- PubMed: 35552369
- DOI: https://doi.org/10.1042/BCJ20220067
- Primary Citation of Related Structures:
7MYJ - PubMed Abstract:
The AMP-activated protein kinase (AMPK) αβγ heterotrimer is a primary cellular energy sensor and central regulator of energy homeostasis. Activating skeletal muscle AMPK with small molecule drugs improves glucose uptake and provides an opportunity for new strategies to treat type 2 diabetes and insulin resistance, with recent genetic and pharmacological studies indicating the α2β2γ1 isoform combination as the heterotrimer complex primarily responsible. With the goal of developing α2β2-specific activators, here we perform structure/function analysis of the 2-hydroxybiphenyl group of SC4, an activator with tendency for α2-selectivity that is also capable of potently activating β2 complexes. Substitution of the LHS 2-hydroxyphenyl group with polar-substituted cyclohexene-based probes resulted in two AMPK agonists, MSG010 and MSG011, which did not display α2-selectivity when screened against a panel of AMPK complexes. By radiolabel kinase assay, MSG010 and MSG011 activated α2β2γ1 AMPK with one order of magnitude greater potency than the pan AMPK activator MK-8722. A crystal structure of MSG011 complexed to AMPK α2β1γ1 revealed a similar binding mode to SC4 and the potential importance of an interaction between the SC4 2-hydroxyl group and α2-Lys31 for directing α2-selectivity. MSG011 induced robust AMPK signalling in mouse primary hepatocytes and commonly used cell lines, and in most cases this occurred in the absence of changes in phosphorylation of the kinase activation loop residue α-Thr172, a classical marker of AMP-induced AMPK activity. These findings will guide future design of α2β2-selective AMPK activators, that we hypothesise may avoid off-target complications associated with indiscriminate activation of AMPK throughout the body.
- Metabolic Signalling Laboratory, St. Vincent's Institute of Medical Research, Fitzroy 3065, Australia.
Organizational Affiliation: