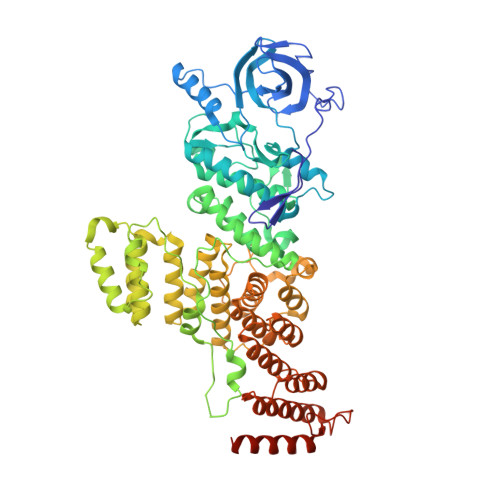
A Tetratricopeptide Repeat Scaffold Couples Signal Detection to OdhI Phosphorylation in Metabolic Control by the Protein Kinase PknG.
Lisa, M.N., Sogues, A., Barilone, N., Baumgart, M., Gil, M., Grana, M., Duran, R., Biondi, R.M., Bellinzoni, M., Bott, M., Alzari, P.M.(2021) mBio 12: e0171721-e0171721
- PubMed: 34607462
- DOI: https://doi.org/10.1128/mBio.01717-21
- Primary Citation of Related Structures:
7MXB, 7MXJ, 7MXK - PubMed Abstract:
Signal transduction is essential for bacteria to adapt to changing environmental conditions. Among many forms of posttranslational modifications, reversible protein phosphorylation has evolved as a ubiquitous molecular mechanism of protein regulation in response to specific stimuli. The Ser/Thr protein kinase PknG modulates the fate of intracellular glutamate by controlling the phosphorylation status of the 2-oxoglutarate dehydrogenase regulator OdhI, a function that is conserved among diverse actinobacteria. PknG has a modular organization characterized by the presence of regulatory domains surrounding the catalytic domain. Here, we present an investigation using in vivo experiments, as well as biochemical and structural methods, of the molecular basis of the regulation of PknG from Corynebacterium glutamicum ( Cg PknG), in the light of previous knowledge available for the kinase from Mycobacterium tuberculosis ( Mtb PknG). We found that OdhI phosphorylation by Cg PknG is regulated by a conserved mechanism that depends on a C-terminal domain composed of tetratricopeptide repeats (TPRs) essential for metabolic homeostasis. Furthermore, we identified a conserved structural motif that physically connects the TPR domain to a β-hairpin within the flexible N-terminal region that is involved in docking interactions with OdhI. Based on our results and previous reports, we propose a model in which the TPR domain of PknG couples signal detection to the specific phosphorylation of OdhI. Overall, the available data indicate that conserved PknG domains in distant actinobacteria retain their roles in kinase regulation in response to nutrient availability. IMPORTANCE Bacteria control the metabolic processes by which they obtain nutrients and energy in order to adapt to the environment. Actinobacteria , one of the largest bacterial phyla of major importance for biotechnology, medicine, and agriculture, developed a unique control process that revolves around a key protein, the protein kinase PknG. Here, we use genetic, biochemical, and structural approaches to study PknG in a system that regulates glutamate production in Corynebacterium glutamicum, a species used for the industrial production of amino acids. The reported findings are conserved in related Actinobacteria , with broader significance for microorganisms that cause disease, as well as environmental species used industrially to produce amino acids and antibiotics every year.
- Unité de Microbiologie Structurale, Institut Pasteurgrid.428999.7grid.418532.9grid.428999.7, CNRS UMR 3528, Université de Paris, Paris, France.
Organizational Affiliation: