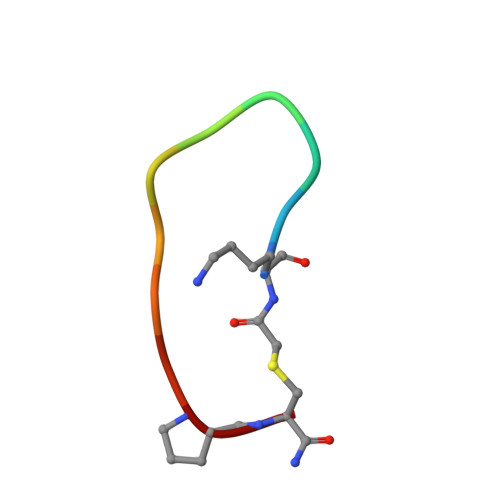
Enhancing the Bioactivity of Bicyclic Peptides Targeted to Grb7-SH2 by Restoring Cell Permeability.
Sturre, N.P., Colson, R.N., Shah, N., Watson, G.M., Yang, X., Wilce, M.C.J., Price, J.T., Wilce, J.A.(2022) Biomedicines 10
- PubMed: 35625882
- DOI: https://doi.org/10.3390/biomedicines10051145
- Primary Citation of Related Structures:
7MP3 - PubMed Abstract:
The development of peptide inhibitors against intracellular targets depends upon the dual challenge of achieving a high affinity and specificity for the target and maintaining cellular permeability for biological activity. Previous efforts to develop bicyclic peptides targeted to the Grb7 signalling protein implicated in HER2+ve cancer progression have resulted in improved affinity. However, these same peptides demonstrated a lowered activity due to their decreased ability to penetrate cell membranes. Here, we report the testing of a new series of bicyclic G7 peptides designed to possess improved bioactivity. We discovered that the incorporation of two amino acids (Phe-Pro, Phe-Trp or Phe-Arg) within the bicyclic peptide framework maintains an enhanced binding affinity for the Grb7-SH2 domain compared to that of the first-generation monocyclic peptide G7-18NATE. Structure determination using X-ray crystallography revealed that the mode of binding by the expanded bicyclic G7 peptide is analogous to that of G7-18NATE. Interestingly, while the bicyclic peptide containing Phe-Trp did not display the highest affinity for Grb7-SH2 in the series, it was the most potent inhibitor of HER2+ve SKBR3 breast cancer cell migration when coupled to Penetratin. Together, this demonstrates that peptide flexibility as well as the amino acid tryptophan can play important roles in the uptake of peptides into the cell.
- Department of Biochemistry and Molecular Biology, Biomedicine Discovery Institute, Monash University, Wellington Road, Clayton, VIC 3800, Australia.
Organizational Affiliation: