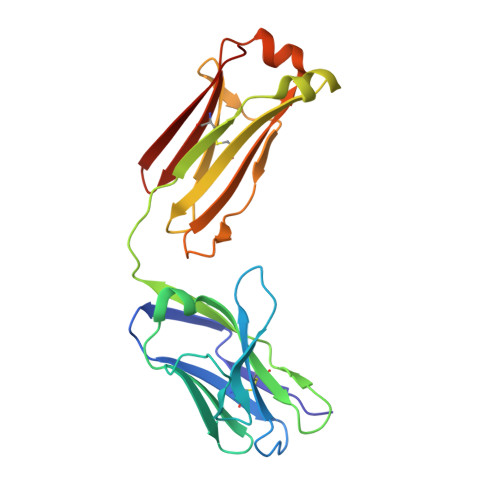
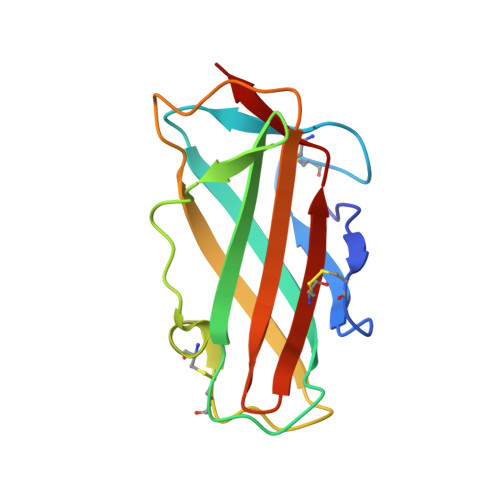
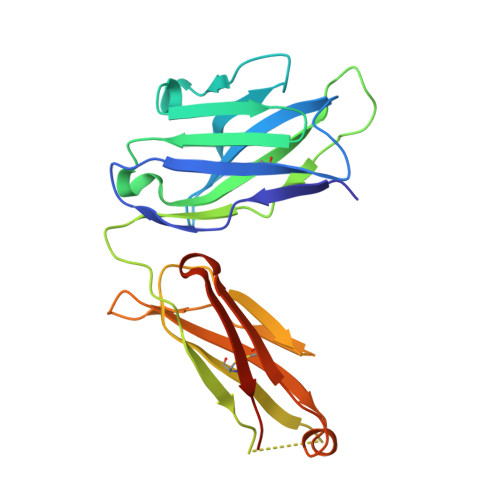
Human IgE monoclonal antibody recognition of mite allergen Der p 2 defines structural basis of an epitope for IgE cross-linking and anaphylaxis in vivo.
Khatri, K., Richardson, C.M., Glesner, J., Kapingidza, A.B., Mueller, G.A., Zhang, J., Dolamore, C., Vailes, L.D., Wunschmann, S., Peebles Jr., R.S., Chapman, M.D., Smith, S.A., Chruszcz, M., Pomes, A.(2022) PNAS Nexus 1: pgac054-pgac054
- PubMed: 35799831
- DOI: https://doi.org/10.1093/pnasnexus/pgac054
- Primary Citation of Related Structures:
7MLH - PubMed Abstract:
Immunoglobulin E (IgE) antibody is a critical effector molecule for adaptive allergen-induced immune responses, which affect up to 40% of the population worldwide. Allergens are usually innocuous molecules but induce IgE antibody production in allergic subjects. Allergen cross-linking of IgE bound to its high affinity receptor (FcεRI) on mast cells and basophils triggers release of histamine and other mediators that cause allergic symptoms. Little is known about the direct allergen-IgE antibody interaction due to the polyclonal nature of serum IgE and the low frequency of IgE-producing B cells in blood. Here, we report the X-ray crystal structure of a house dust mite allergen, Der p 2, in complex with Fab of a human IgE monoclonal antibody (mAb) isolated by hybridoma technology using human B cells from an allergic subject. This IgE mAb, 2F10, has the correct pairing of heavy and light chains as it occurs in vivo . Key amino acids forming the IgE epitope on Der p 2 were identified. Mutation of these residues ablated their functional ability to cross-link IgE in a mouse model of passive systemic anaphylaxis. These analyses revealed an important conformational epitope associated with the IgE antibody repertoire to a major mite allergen.
- Department of Chemistry and Biochemistry, University of South Carolina, Columbia, SC 29208, USA.
Organizational Affiliation: