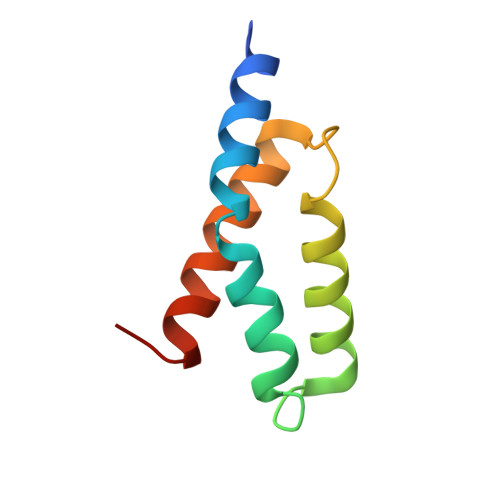
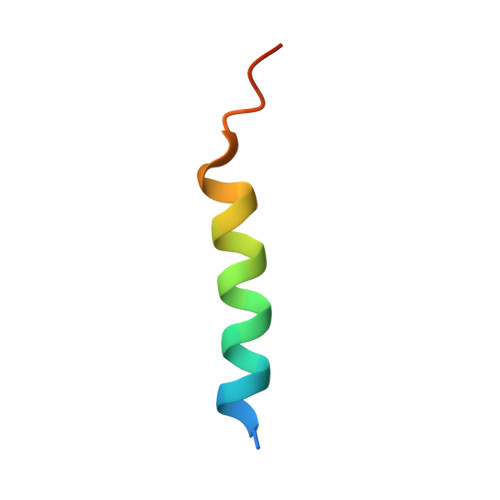
Panoramix SUMOylation on chromatin connects the piRNA pathway to the cellular heterochromatin machinery.
Andreev, V.I., Yu, C., Wang, J., Schnabl, J., Tirian, L., Gehre, M., Handler, D., Duchek, P., Novatchkova, M., Baumgartner, L., Meixner, K., Sienski, G., Patel, D.J., Brennecke, J.(2022) Nat Struct Mol Biol 29: 130-142
- PubMed: 35173350
- DOI: https://doi.org/10.1038/s41594-022-00721-x
- Primary Citation of Related Structures:
7MKK - PubMed Abstract:
Nuclear Argonaute proteins, guided by small RNAs, mediate sequence-specific heterochromatin formation. The molecular principles that link Argonaute-small RNA complexes to cellular heterochromatin effectors on binding to nascent target RNAs are poorly understood. Here, we explain the mechanism by which the PIWI-interacting RNA (piRNA) pathway connects to the heterochromatin machinery in Drosophila. We find that Panoramix, a corepressor required for piRNA-guided heterochromatin formation, is SUMOylated on chromatin in a Piwi-dependent manner. SUMOylation, together with an amphipathic LxxLL motif in Panoramix's intrinsically disordered repressor domain, are necessary and sufficient to recruit Small ovary (Sov), a multi-zinc-finger protein essential for general heterochromatin formation and viability. Structure-guided mutations that eliminate the Panoramix-Sov interaction or that prevent SUMOylation of Panoramix uncouple Sov from the piRNA pathway, resulting in viable but sterile flies in which Piwi-targeted transposons are derepressed. Thus, Piwi engages the heterochromatin machinery specifically at transposon loci by coupling recruitment of a corepressor to nascent transcripts with its SUMOylation.
- Institute of Molecular Biotechnology of the Austrian Academy of Sciences (IMBA), Vienna BioCenter (VBC), Vienna, Austria.
Organizational Affiliation: