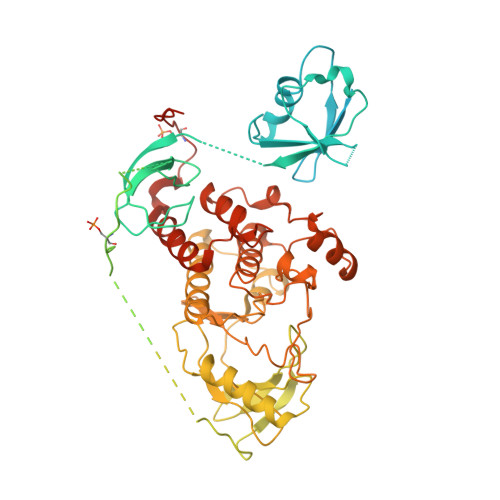
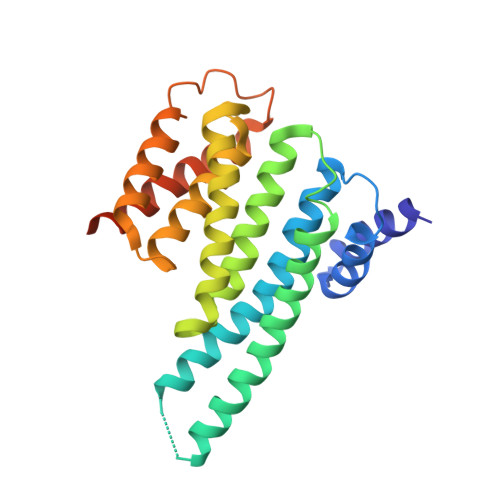
Structural insights into the BRAF monomer-to-dimer transition mediated by RAS binding.
Martinez Fiesco, J.A., Durrant, D.E., Morrison, D.K., Zhang, P.(2022) Nat Commun 13: 486-486
- PubMed: 35078985
- DOI: https://doi.org/10.1038/s41467-022-28084-3
- Primary Citation of Related Structures:
7MFD, 7MFE, 7MFF - PubMed Abstract:
RAF kinases are essential effectors of RAS, but how RAS binding initiates the conformational changes needed for autoinhibited RAF monomers to form active dimers has remained unclear. Here, we present cryo-electron microscopy structures of full-length BRAF complexes derived from mammalian cells: autoinhibited, monomeric BRAF:14-3-3 2 :MEK and BRAF:14-3-3 2 complexes, and an inhibitor-bound, dimeric BRAF 2 :14-3-3 2 complex, at 3.7, 4.1, and 3.9 Å resolution, respectively. In both autoinhibited, monomeric structures, the RAS binding domain (RBD) of BRAF is resolved, revealing that the RBD forms an extensive contact interface with the 14-3-3 protomer bound to the BRAF C-terminal site and that key basic residues required for RBD-RAS binding are exposed. Moreover, through structure-guided mutational studies, our findings indicate that RAS-RAF binding is a dynamic process and that RBD residues at the center of the RBD:14-3-3 interface have a dual function, first contributing to RAF autoinhibition and then to the full spectrum of RAS-RBD interactions.
- Center for Structural Biology, Center for Cancer Research, National Cancer Institute-Frederick, Frederick, MD, 21702, USA.
Organizational Affiliation: