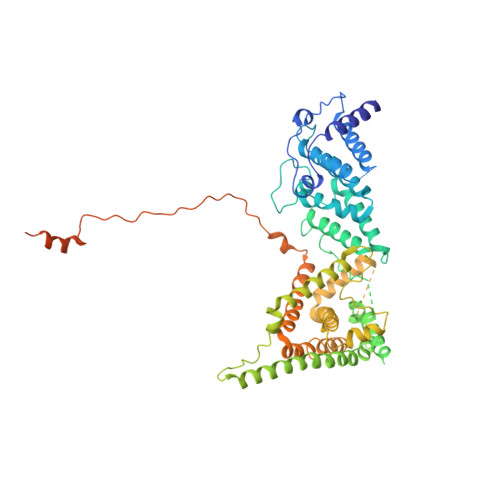
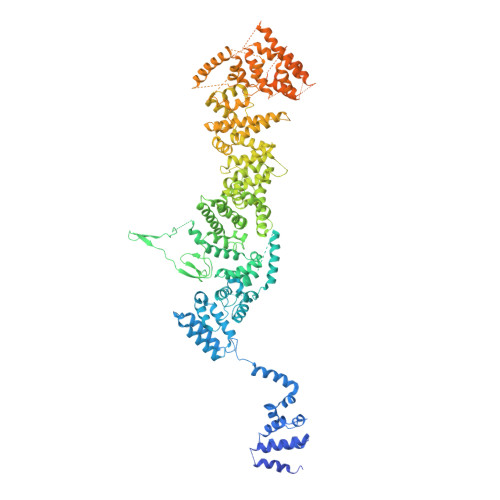
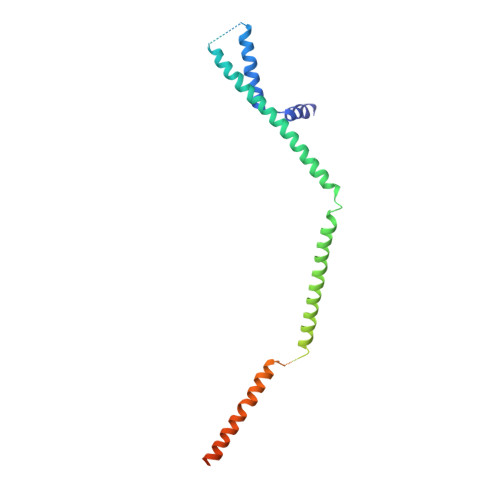
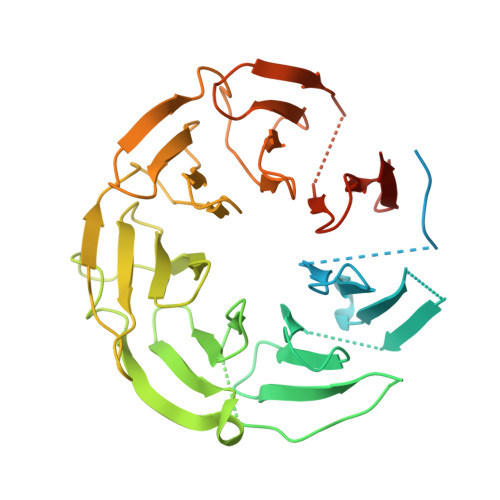
Cryo-EM structure of the yeast TREX complex and coordination with the SR-like protein Gbp2.
Xie, Y., Clarke, B.P., Kim, Y.J., Ivey, A.L., Hill, P.S., Shi, Y., Ren, Y.(2021) Elife 10
- PubMed: 33787496
- DOI: https://doi.org/10.7554/eLife.65699
- Primary Citation of Related Structures:
7LUV - PubMed Abstract:
The evolutionarily conserved TRanscript-EXport (TREX) complex plays central roles during mRNP (messenger ribonucleoprotein) maturation and export from the nucleus to the cytoplasm. In yeast, TREX is composed of the THO sub-complex (Tho2, Hpr1, Tex1, Mft1, and Thp2), the DEAD box ATPase Sub2, and Yra1. Here we present a 3.7 Å cryo-EM structure of the yeast THO•Sub2 complex. The structure reveals the intimate assembly of THO revolving around its largest subunit Tho2. THO stabilizes a semi-open conformation of the Sub2 ATPase via interactions with Tho2. We show that THO interacts with the serine-arginine (SR)-like protein Gbp2 through both the RS domain and RRM domains of Gbp2. Cross-linking mass spectrometry analysis supports the extensive interactions between THO and Gbp2, further revealing that RRM domains of Gbp2 are in close proximity to the C-terminal domain of Tho2. We propose that THO serves as a landing pad to configure Gbp2 to facilitate its loading onto mRNP.
- Department of Biochemistry, Vanderbilt University School of Medicine, Nashville, United States.
Organizational Affiliation: