Discovery and Optimization of DNA Gyrase and Topoisomerase IV Inhibitors with Potent Activity against Fluoroquinolone-Resistant Gram-Positive Bacteria.
Lapointe, G., Skepper, C.K., Holder, L.M., Armstrong, D., Bellamacina, C., Blais, J., Bussiere, D., Bian, J., Cepura, C., Chan, H., Dean, C.R., De Pascale, G., Dhumale, B., Fisher, L.M., Fulsunder, M., Kantariya, B., Kim, J., King, S., Kossy, L., Kulkarni, U., Lakshman, J., Leeds, J.A., Ling, X., Lvov, A., Ma, S., Malekar, S., McKenney, D., Mergo, W., Metzger, L., Mhaske, K., Moser, H.E., Mostafavi, M., Namballa, S., Noeske, J., Osborne, C., Patel, A., Patel, D., Patel, T., Piechon, P., Polyakov, V., Prajapati, K., Prosen, K.R., Reck, F., Richie, D.L., Sanderson, M.R., Satasia, S., Savani, B., Selvarajah, J., Sethuraman, V., Shu, W., Tashiro, K., Thompson, K.V., Vaarla, K., Vala, L., Veselkov, D.A., Vo, J., Vora, B., Wagner, T., Wedel, L., Williams, S.L., Yendluri, S., Yue, Q., Yifru, A., Zhang, Y., Rivkin, A.(2021) J Med Chem 64: 6329-6357
- PubMed: 33929852
- DOI: https://doi.org/10.1021/acs.jmedchem.1c00375
- Primary Citation of Related Structures:
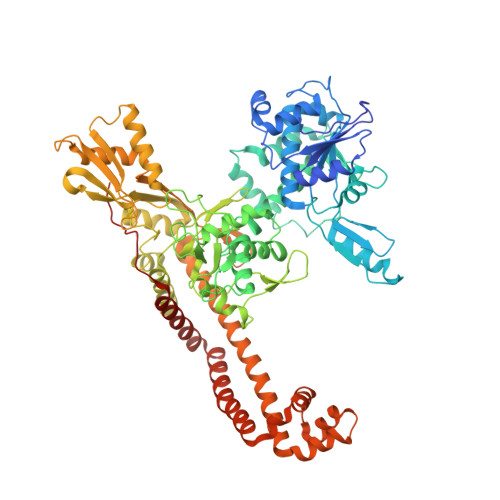
7LHZ - PubMed Abstract:
Herein, we describe the discovery and optimization of a novel series that inhibits bacterial DNA gyrase and topoisomerase IV via binding to, and stabilization of, DNA cleavage complexes. Optimization of this series led to the identification of compound 25 , which has potent activity against Gram-positive bacteria, a favorable in vitro safety profile, and excellent in vivo pharmacokinetic properties. Compound 25 was found to be efficacious against fluoroquinolone-sensitive Staphylococcus aureus infection in a mouse thigh model at lower doses than moxifloxacin. An X-ray crystal structure of the ternary complex formed by topoisomerase IV from Klebsiella pneumoniae , compound 25 , and cleaved DNA indicates that this compound does not engage in a water-metal ion bridge interaction and forms no direct contacts with residues in the quinolone resistance determining region (QRDR). This suggests a structural basis for the reduced impact of QRDR mutations on antibacterial activity of 25 compared to fluoroquinolones.
- Novartis Institutes for BioMedical Research, Emeryville, California 94608, United States.
Organizational Affiliation: