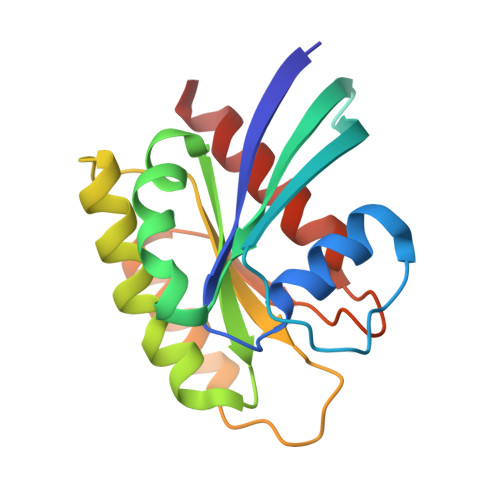
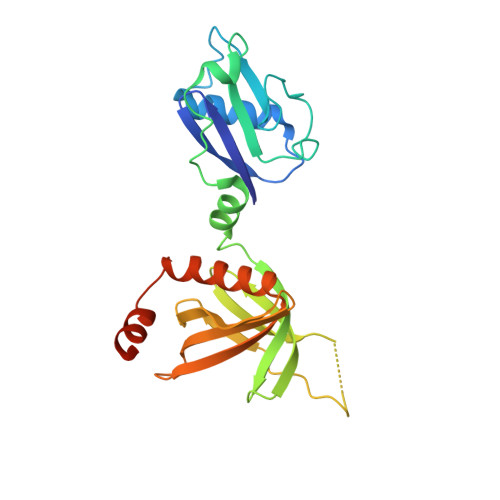
RAS interaction with Sin1 is dispensable for mTORC2 assembly and activity.
Castel, P., Dharmaiah, S., Sale, M.J., Messing, S., Rizzuto, G., Cuevas-Navarro, A., Cheng, A., Trnka, M.J., Urisman, A., Esposito, D., Simanshu, D.K., McCormick, F.(2021) Proc Natl Acad Sci U S A 118
- PubMed: 34380736
- DOI: https://doi.org/10.1073/pnas.2103261118
- Primary Citation of Related Structures:
7LC1, 7LC2 - PubMed Abstract:
RAS proteins are molecular switches that interact with effector proteins when bound to guanosine triphosphate, stimulating downstream signaling in response to multiple stimuli. Although several canonical downstream effectors have been extensively studied and tested as potential targets for RAS-driven cancers, many of these remain poorly characterized. In this study, we undertook a biochemical and structural approach to further study the role of Sin1 as a RAS effector. Sin1 interacted predominantly with KRAS isoform 4A in cells through an atypical RAS-binding domain that we have characterized by X-ray crystallography. Despite the essential role of Sin1 in the assembly and activity of mTORC2, we find that the interaction with RAS is not required for these functions. Cells and mice expressing a mutant of Sin1 that is unable to bind RAS are proficient for activation and assembly of mTORC2. Our results suggest that Sin1 is a bona fide RAS effector that regulates downstream signaling in an mTORC2-independent manner.
- Helen Diller Family Comprehensive Cancer Center, University of California, San Francisco, CA 94158.
Organizational Affiliation: