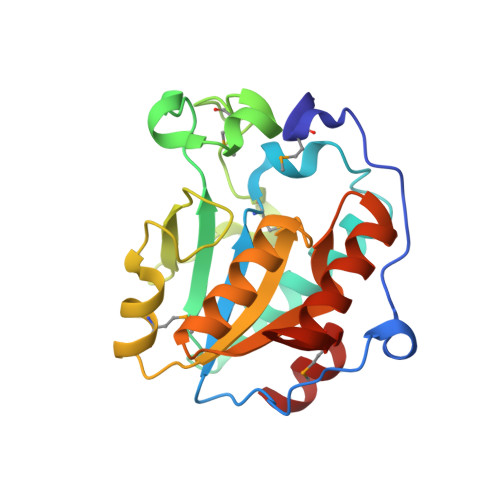
Functional and structural characterization of Stenotrophomonas maltophilia EntB, an unusual form of isochorismatase for siderophore synthesis.
Nas, M.Y., Gabell, J., Inniss, N., Minasov, G., Shuvalova, L., Satchell, K.J.F., Cianciotto, N.P.(2025) Acta Crystallogr F Struct Biol Commun 81: 287-296
- PubMed: 40464617
- DOI: https://doi.org/10.1107/S2053230X2500490X
- Primary Citation of Related Structures:
7L6J - PubMed Abstract:
Clinical and environmental isolates of Stenotrophomonas maltophilia produce an enterobactin-like siderophore that promotes bacterial growth under low-iron conditions. Although prior mutational and bioinformatic analyses indicated that most of the enzymes encoded by the S. maltophilia entCEBB'FA locus are suitably reminiscent of their counterparts in Escherichia coli and other bacteria, Stenotrophomonas EntB was unusual. In bacteria producing enterobactin-related molecules, EntB and its homologs are usually multi-domain proteins in which the amino portion acts as an isochorismatase and the carboxy domain serves as an aryl carrier protein (ArCP). However, in S. maltophilia the isochorismatase and ArCP functions are encoded by two distinct genes: entB and entB', respectively. Current mutant analysis was used to first confirm that S. maltophilia entB is needed for siderophore activity and bacterial growth in iron-depleted media. A crystal structure of S. maltophilia EntB was then obtained. The structure aligned with the N-terminal portion of EntB from E. coli and VibB from Vibrio cholerae, affirming the protein to be a single-domain isochorismatase. However, S. maltophilia EntB also aligned with the single-domain PhzD from Pseudomonas aeruginosa, which is a key enzyme involved in the biosynthesis of the antimicrobial compound phenazine. BLASTP searches indicated that entB and its neighboring genes are fully conserved amongst S. maltophilia strains but are variably present in other Stenotrophomonas species. The closest homologs to S. maltophilia EntB outside the genus were hypothetical proteins/putative isochorismatases in some Gram-negative bacteria (for example Pseudomonas spp. and Xanthomonas spp.), Gram-positive bacteria (Streptomyces spp. and Bacillus subtilis) and fungi (for example Rhizopus arrhizus and Knufia peltigerae).
- Department of Microbiology-Immunology, Northwestern University Feinberg School of Medicine, Chicago, IL 60611, USA.
Organizational Affiliation: