The internal aldimine form of the wild-type Salmonella typhimurium Tryptophan Synthase with cesium ion at the metal coordination site at 1.60 Angstrom resolution
Hilario, E., Dunn, M.F., Mueller, L.J.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Tryptophan synthase alpha chain | 268 | Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 | Mutation(s): 0 Gene Names: trpA, STM1727 EC: 4.2.1.20 | 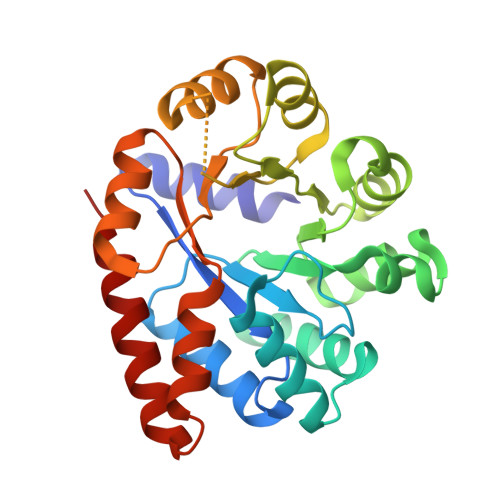 | |
UniProt | |||||
Find proteins for P00929 (Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720)) Explore P00929 Go to UniProtKB: P00929 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P00929 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Tryptophan synthase beta chain | 397 | Salmonella enterica subsp. enterica serovar Typhimurium | Mutation(s): 0 Gene Names: trpB EC: 4.2.1.20 | 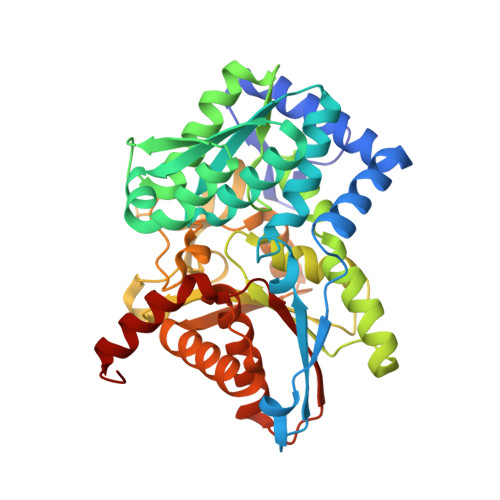 | |
UniProt | |||||
Find proteins for P0A2K1 (Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720)) Explore P0A2K1 Go to UniProtKB: P0A2K1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A2K1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 7 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| PLP (Subject of Investigation/LOI) Query on PLP | W [auth B] | PYRIDOXAL-5'-PHOSPHATE C8 H10 N O6 P NGVDGCNFYWLIFO-UHFFFAOYSA-N |  | ||
| CS (Subject of Investigation/LOI) Query on CS | GA [auth B], U [auth A], YA [auth B] | CESIUM ION Cs NCMHKCKGHRPLCM-UHFFFAOYSA-N |  | ||
| PEG Query on PEG | JA [auth B], SA [auth B] | DI(HYDROXYETHYL)ETHER C4 H10 O3 MTHSVFCYNBDYFN-UHFFFAOYSA-N |  | ||
| GOL Query on GOL | J [auth A] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| DMS Query on DMS | AA [auth B] BA [auth B] CA [auth B] D [auth A] DA [auth B] | DIMETHYL SULFOXIDE C2 H6 O S IAZDPXIOMUYVGZ-UHFFFAOYSA-N |  | ||
| EDO Query on EDO | C [auth A] KA [auth B] L [auth A] MA [auth B] O [auth A] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| CL Query on CL | HA [auth B] IA [auth B] S [auth A] T [auth A] VA [auth B] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 182.549 | α = 90 |
| b = 59.188 | β = 94.48 |
| c = 67.073 | γ = 90 |
| Software Name | Purpose |
|---|---|
| xia2 | data reduction |
| SCALA | data scaling |
| MOLREP | phasing |
| DM | phasing |
| REFMAC | refinement |
| PDB_EXTRACT | data extraction |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Human Genome Research Institute (NIH/NHGRI) | United States | 2R01GM097569-06A1 |