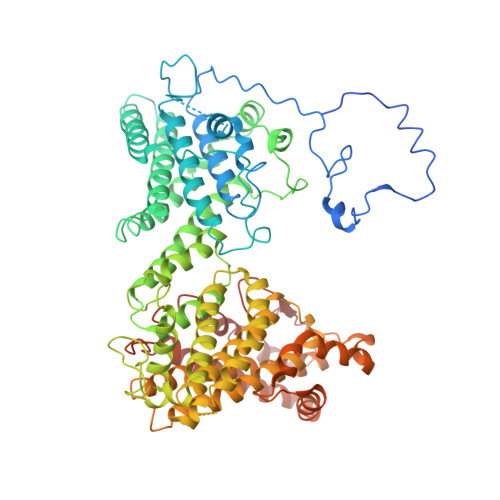
Molecular mechanism of N-terminal acetylation by the ternary NatC complex.
Deng, S., Gottlieb, L., Pan, B., Supplee, J., Wei, X., Petersson, E.J., Marmorstein, R.(2021) Structure 29: 1094
- PubMed: 34019809
- DOI: https://doi.org/10.1016/j.str.2021.05.003
- Primary Citation of Related Structures:
7L1K - PubMed Abstract:
Protein N-terminal acetylation is predominantly a ribosome-associated modification, with NatA-E serving as the major enzymes. NatC is the most unusual of these enzymes, containing one Naa30 catalytic subunit and two auxiliary subunits, Naa35 and Naa38; and substrate selectivity profile that overlaps with NatE. Here, we report the cryoelectron microscopy structure of S. pombe NatC with a NatE/C-type bisubstrate analog and inositol hexaphosphate (IP 6 ), and associated biochemistry studies. We find that the presence of three subunits is a prerequisite for normal NatC acetylation activity in yeast and that IP 6 binds tightly to NatC to stabilize the complex. We also describe the molecular basis for IP 6 -mediated NatC complex stabilization and the overlapping yet distinct substrate profiles of NatC and NatE.
- Department of Chemistry, 231 South 34(th) Street, University of Pennsylvania, Philadelphia, PA 19104, USA; Abramson Family Cancer Research Institute, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA.
Organizational Affiliation: