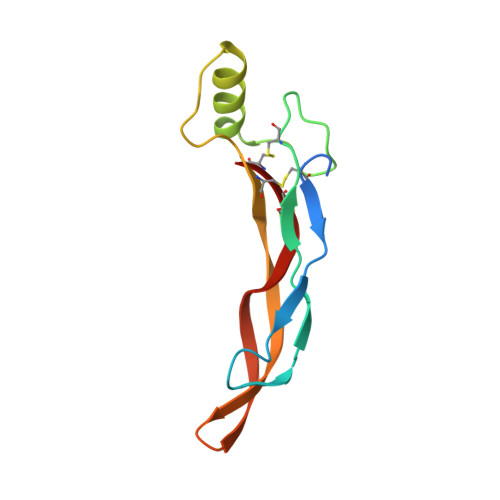
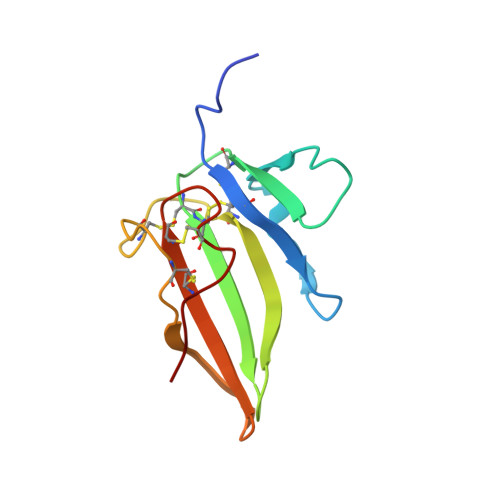
Structure of AMH bound to AMHR2 provides insight into a unique signaling pair in the TGF-beta family.
Hart, K.N., Stocker, W.A., Nagykery, N.G., Walton, K.L., Harrison, C.A., Donahoe, P.K., Pepin, D., Thompson, T.B.(2021) Proc Natl Acad Sci U S A 118
- PubMed: 34155118
- DOI: https://doi.org/10.1073/pnas.2104809118
- Primary Citation of Related Structures:
7L0J - PubMed Abstract:
Anti-Müllerian hormone (AMH), or Müllerian-inhibiting substance, is a protein hormone that promotes Müllerian duct regression during male fetal sexual differentiation and regulation of folliculogenesis in women. AMH is a member of the transforming growth factor beta (TGF-β) family, which has evolved to signal through its own dedicated type II receptor, AMH receptor type II (AMHR2). Structures of other TGF-β family members have revealed how ligands infer specificity for their cognate receptors; however, it is unknown how AMH binds AMHR2 at the molecular level. Therefore, in this study, we solved the X-ray crystal structure of AMH bound to the extracellular domain of AMHR2 to a resolution of 2.6Å. The structure reveals that while AMH binds AMHR2 in a similar location to Activin and BMP ligand binding to their type II receptors, differences in both AMH and AMHR2 account for a highly specific interaction. Furthermore, using an AMH responsive cell-based luciferase assay, we show that a conformation in finger 1 of AMHR2 and a salt bridge formed by K534 on AMH and D81/E84 of AMHR2 are key to the AMH/AMHR2 interaction. Overall, our study highlights how AMH engages AMHR2 using a modified paradigm of receptor binding facilitated by modifications to the three-finger toxin fold of AMHR2. Furthermore, understanding these elements contributing to the specificity of binding will help in the design of agonists or antagonists or the selection of antibody therapies.
- Department of Pharmacology and Systems Physiology, University of Cincinnati, Cincinnati, OH 45267.
Organizational Affiliation: