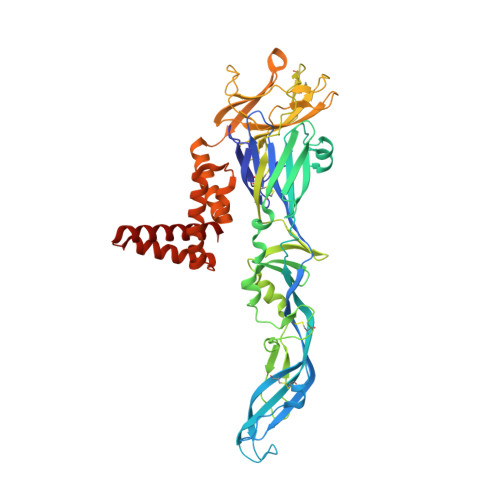
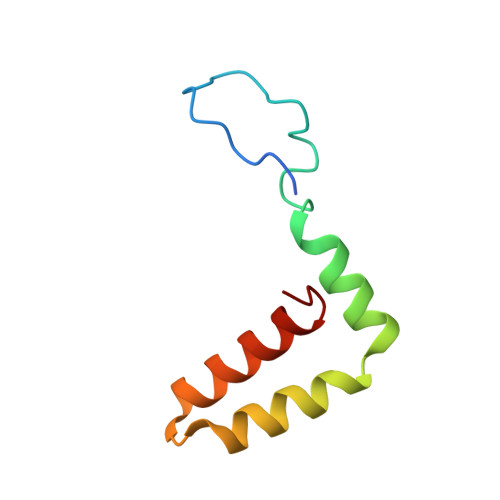
A unified route for flavivirus structures uncovers essential pocket factors conserved across pathogenic viruses.
Hardy, J.M., Newton, N.D., Modhiran, N., Scott, C.A.P., Venugopal, H., Vet, L.J., Young, P.R., Hall, R.A., Hobson-Peters, J., Coulibaly, F., Watterson, D.(2021) Nat Commun 12: 3266-3266
- PubMed: 34075032
- DOI: https://doi.org/10.1038/s41467-021-22773-1
- Primary Citation of Related Structures:
7KV8, 7KV9, 7KVA, 7KVB - PubMed Abstract:
The epidemic emergence of relatively rare and geographically isolated flaviviruses adds to the ongoing disease burden of viruses such as dengue. Structural analysis is key to understand and combat these pathogens. Here, we present a chimeric platform based on an insect-specific flavivirus for the safe and rapid structural analysis of pathogenic viruses. We use this approach to resolve the architecture of two neurotropic viruses and a structure of dengue virus at 2.5 Å, the highest resolution for an enveloped virion. These reconstructions allow improved modelling of the stem region of the envelope protein, revealing two lipid-like ligands within highly conserved pockets. We show that these sites are essential for viral growth and important for viral maturation. These findings define a hallmark of flavivirus virions and a potential target for broad-spectrum antivirals and vaccine design. We anticipate the chimeric platform to be widely applicable for investigating flavivirus biology.
- Infection and Immunity Program, Biomedicine Discovery Institute and Department of Biochemistry and Molecular Biology, Monash University, Clayton, VIC, Australia.
Organizational Affiliation: