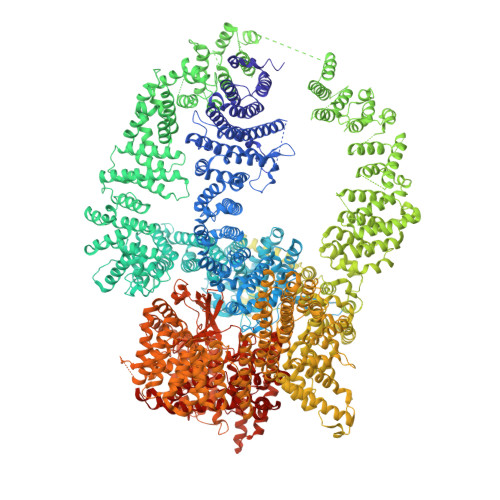
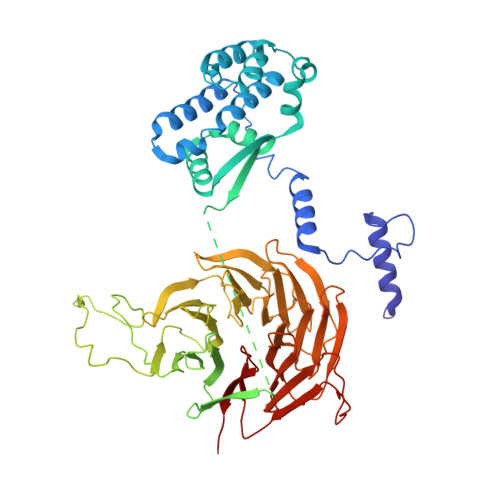
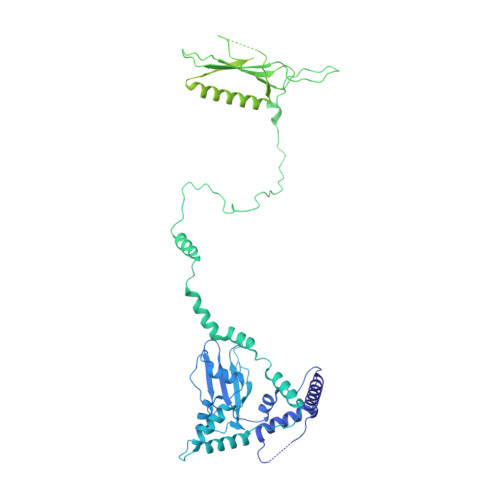
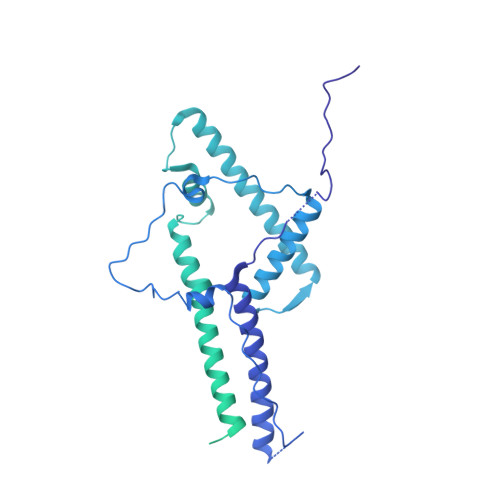
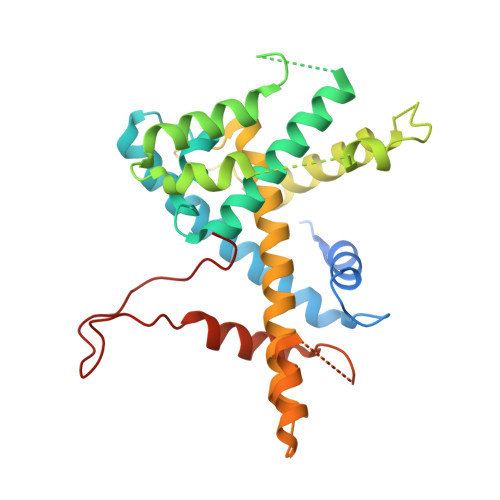
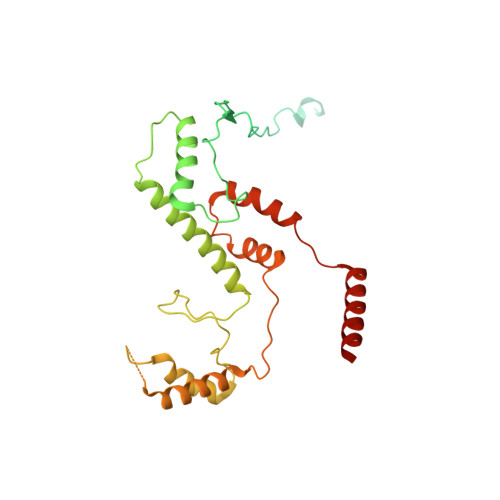
Structure of the human SAGA coactivator complex.
Herbst, D.A., Esbin, M.N., Louder, R.K., Dugast-Darzacq, C., Dailey, G.M., Fang, Q., Darzacq, X., Tjian, R., Nogales, E.(2021) Nat Struct Mol Biol 28: 989-996
- PubMed: 34811519
- DOI: https://doi.org/10.1038/s41594-021-00682-7
- Primary Citation of Related Structures:
7KTR, 7KTS - PubMed Abstract:
The SAGA complex is a regulatory hub involved in gene regulation, chromatin modification, DNA damage repair and signaling. While structures of yeast SAGA (ySAGA) have been reported, there are noteworthy functional and compositional differences for this complex in metazoans. Here we present the cryogenic-electron microscopy (cryo-EM) structure of human SAGA (hSAGA) and show how the arrangement of distinct structural elements results in a globally divergent organization from that of yeast, with a different interface tethering the core module to the TRRAP subunit, resulting in a dramatically altered geometry of functional elements and with the integration of a metazoan-specific splicing module. Our hSAGA structure reveals the presence of an inositol hexakisphosphate (InsP 6 ) binding site in TRRAP and an unusual property of its pseudo-(Ψ)PIKK. Finally, we map human disease mutations, thus providing the needed framework for structure-guided drug design of this important therapeutic target for human developmental diseases and cancer.
- California Institute for Quantitative Biology (QB3), University of California, Berkeley, CA, USA.
Organizational Affiliation: