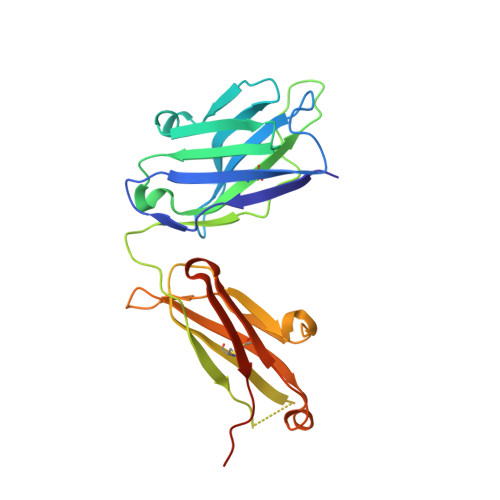
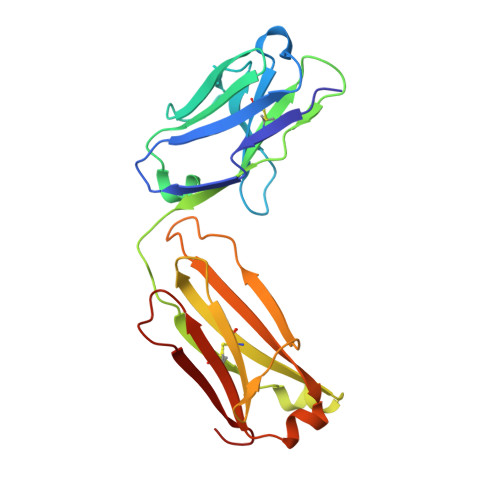
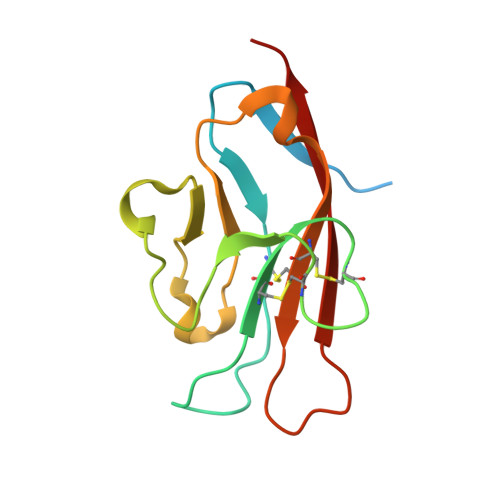
Tim-3 mediates T cell trogocytosis to limit antitumor immunity.
Pagliano, O., Morrison, R.M., Chauvin, J.M., Banerjee, H., Davar, D., Ding, Q., Tanegashima, T., Gao, W., Chakka, S.R., DeBlasio, R., Lowin, A., Kara, K., Ka, M., Zidi, B., Amin, R., Raphael, I., Zhang, S., Watkins, S.C., Sander, C., Kirkwood, J.M., Bosenberg, M., Anderson, A.C., Kuchroo, V.K., Kane, L.P., Korman, A.J., Rajpal, A., West, S.M., Han, M., Bee, C., Deng, X., Schebye, X.M., Strop, P., Zarour, H.M.(2022) J Clin Invest 132
- PubMed: 35316223
- DOI: https://doi.org/10.1172/JCI152864
- Primary Citation of Related Structures:
7KQL - PubMed Abstract:
T cell immunoglobulin mucin domain-containing protein 3 (Tim-3) negatively regulates innate and adaptive immunity in cancer. To identify the mechanisms of Tim-3 in cancer immunity, we evaluated the effects of Tim-3 blockade in human and mouse melanoma. Here, we show that human programmed cell death 1-positive (PD-1+) Tim-3+CD8+ tumor-infiltrating lymphocytes (TILs) upregulate phosphatidylserine (PS), a receptor for Tim-3, and acquire cell surface myeloid markers from antigen-presenting cells (APCs) through transfer of membrane fragments called trogocytosis. Tim-3 blockade acted on Tim-3+ APCs in a PS-dependent fashion to disrupt the trogocytosis of activated tumor antigen-specific CD8+ T cells and PD-1+Tim-3+ CD8+ TILs isolated from patients with melanoma. Tim-3 and PD-1 blockades cooperated to disrupt trogocytosis of CD8+ TILs in 2 melanoma mouse models, decreasing tumor burden and prolonging survival. Deleting Tim-3 in dendritic cells but not in CD8+ T cells impeded the trogocytosis of CD8+ TILs in vivo. Trogocytosed CD8+ T cells presented tumor peptide-major histocompatibility complexes and became the target of fratricide T cell killing, which was reversed by Tim-3 blockade. Our findings have uncovered a mechanism Tim-3 uses to limit antitumor immunity.
- Department of Medicine and UPMC Hillman Cancer Center.
Organizational Affiliation: