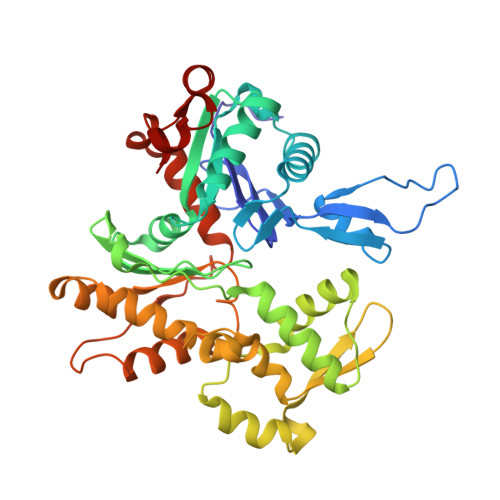
The structure of the native cardiac thin filament at systolic Ca 2+ levels.
Risi, C.M., Pepper, I., Belknap, B., Landim-Vieira, M., White, H.D., Dryden, K., Pinto, J.R., Chase, P.B., Galkin, V.E.(2021) Proc Natl Acad Sci U S A 118
- PubMed: 33753506
- DOI: https://doi.org/10.1073/pnas.2024288118
- Primary Citation of Related Structures:
7KO4, 7KO5, 7KO7, 7KON, 7KOR - PubMed Abstract:
Every heartbeat relies on cyclical interactions between myosin thick and actin thin filaments orchestrated by rising and falling Ca 2+ levels. Thin filaments are comprised of two actin strands, each harboring equally separated troponin complexes, which bind Ca 2+ to move tropomyosin cables away from the myosin binding sites and, thus, activate systolic contraction. Recently, structures of thin filaments obtained at low (pCa ∼9) or high (pCa ∼3) Ca 2+ levels revealed the transition between the Ca 2+ -free and Ca 2+ -bound states. However, in working cardiac muscle, Ca 2+ levels fluctuate at intermediate values between pCa ∼6 and pCa ∼7. The structure of the thin filament at physiological Ca 2+ levels is unknown. We used cryoelectron microscopy and statistical analysis to reveal the structure of the cardiac thin filament at systolic pCa = 5.8. We show that the two strands of the thin filament consist of a mixture of regulatory units, which are composed of Ca 2+ -free, Ca 2+ -bound, or mixed (e.g., Ca 2+ free on one side and Ca 2+ bound on the other side) troponin complexes. We traced troponin complex conformations along and across individual thin filaments to directly determine the structural composition of the cardiac native thin filament at systolic Ca 2+ levels. We demonstrate that the two thin filament strands are activated stochastically with short-range cooperativity evident only on one of the two strands. Our findings suggest a mechanism by which cardiac muscle is regulated by narrow range Ca 2+ fluctuations.
- Department of Physiological Sciences, Eastern Virginia Medical School, Norfolk, VA 23507.
Organizational Affiliation: