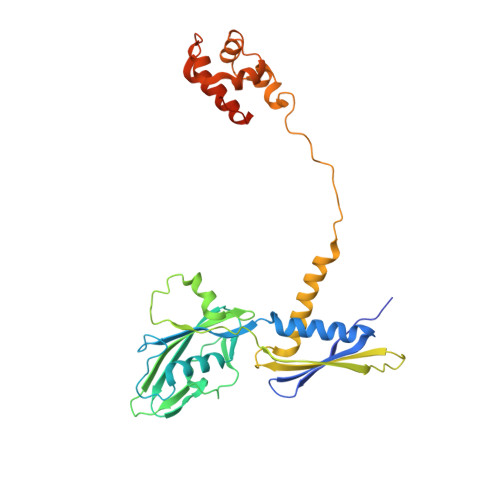
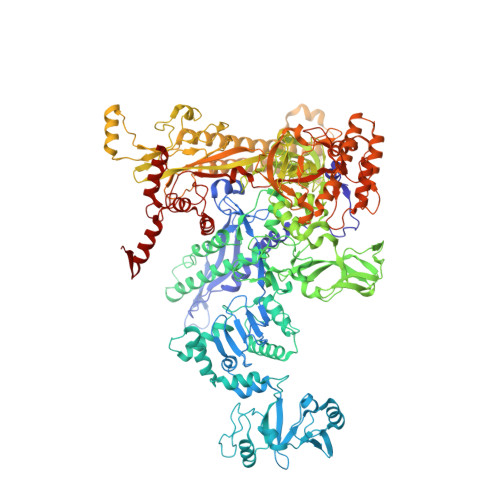
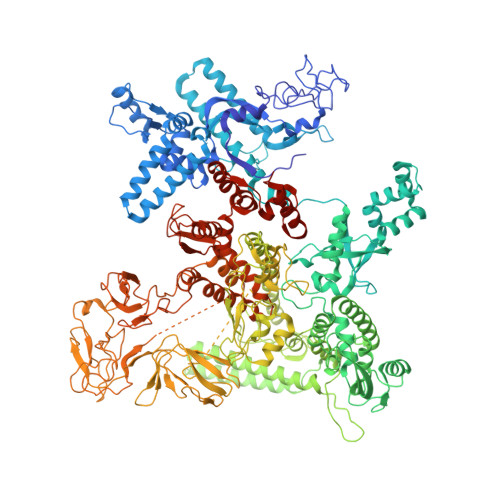
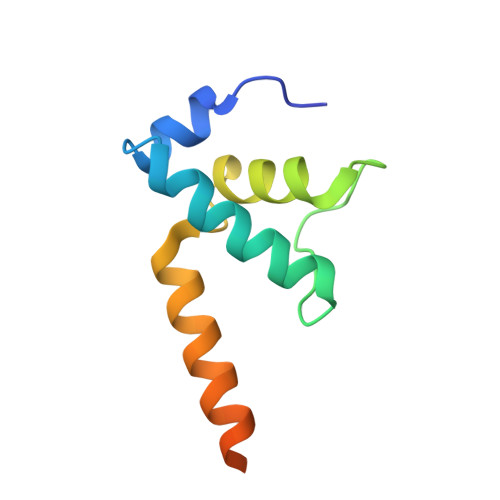
Structural basis of ribosomal RNA transcription regulation.
Shin, Y., Qayyum, M.Z., Pupov, D., Esyunina, D., Kulbachinskiy, A., Murakami, K.S.(2021) Nat Commun 12: 528-528
- PubMed: 33483500
- DOI: https://doi.org/10.1038/s41467-020-20776-y
- Primary Citation of Related Structures:
7KHB, 7KHC, 7KHE, 7KHI - PubMed Abstract:
Ribosomal RNA (rRNA) is most highly expressed in rapidly growing bacteria and is drastically downregulated under stress conditions by the global transcriptional regulator DksA and the alarmone ppGpp. Here, we determined cryo-electron microscopy structures of the Escherichia coli RNA polymerase (RNAP) σ 70 holoenzyme during rRNA promoter recognition with and without DksA/ppGpp. RNAP contacts the UP element using dimerized α subunit carboxyl-terminal domains and scrunches the template DNA with the σ finger and β' lid to select the transcription start site favorable for rapid promoter escape. Promoter binding induces conformational change of σ domain 2 that opens a gate for DNA loading and ejects σ 1.1 from the RNAP cleft to facilitate open complex formation. DksA/ppGpp binding also opens the DNA loading gate, which is not coupled to σ 1.1 ejection and impedes open complex formation. These results provide a molecular basis for the exceptionally active rRNA transcription and its vulnerability to DksA/ppGpp.
- Department of Biochemistry and Molecular Biology, Pennsylvania State University, University Park, PA, 16802, USA.
Organizational Affiliation: