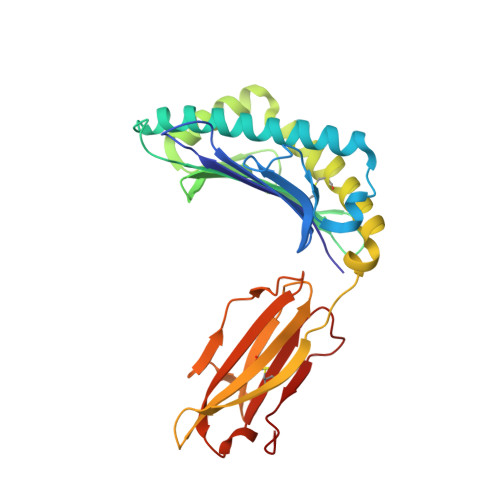
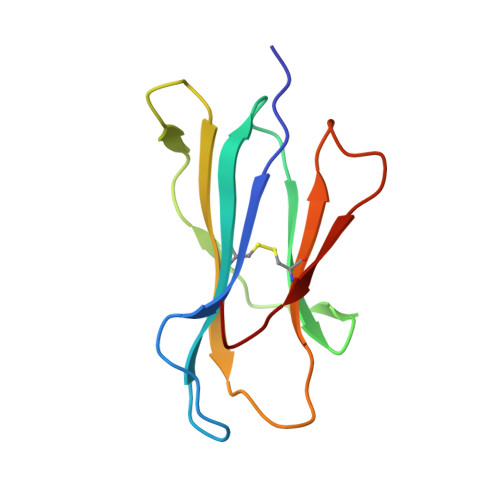
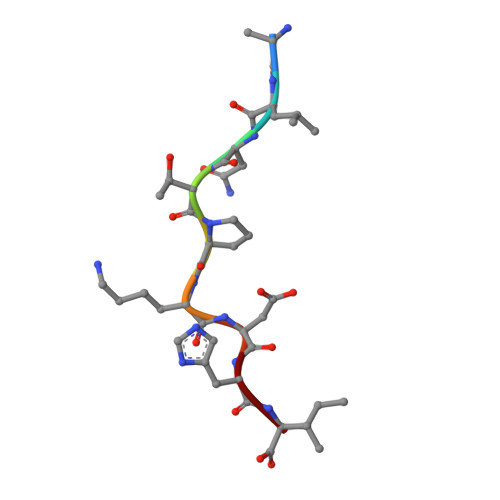
The presentation of SARS-CoV-2 peptides by the common HLA-A*02:01 molecule.
Szeto, C., Chatzileontiadou, D.S.M., Nguyen, A.T., Sloane, H., Lobos, C.A., Jayasinghe, D., Halim, H., Smith, C., Riboldi-Tunnicliffe, A., Grant, E.J., Gras, S.(2021) iScience 24: 102096-102096
- PubMed: 33521593
- DOI: https://doi.org/10.1016/j.isci.2021.102096
- Primary Citation of Related Structures:
7KGO, 7KGP, 7KGQ, 7KGR, 7KGS, 7KGT - PubMed Abstract:
CD8+ T cells are crucial for anti-viral immunity; however, understanding T cell responses requires the identification of epitopes presented by human leukocyte antigens (HLA). To date, few SARS-CoV-2-specific CD8+ T cell epitopes have been described. Internal viral proteins are typically more conserved than surface proteins and are often the target of CD8+ T cells. Therefore, we have characterized eight peptides derived from the internal SARS-CoV-2 nucleocapsid protein predicted to bind HLA-A ∗ 02:01, the most common HLA molecule in the global population. We determined not all peptides could form a complex with HLA-A ∗ 02:01, and the six crystal structures determined revealed that some peptides adopted a mobile conformation. We therefore provide a molecular understanding of SARS-CoV-2 CD8+ T cell epitopes. Furthermore, we show that there is limited pre-existing CD8+ T cell response toward these epitopes in unexposed individuals. Together, these data show that SARS-CoV-2 nucleocapsid might not contain potent epitopes restricted to HLA-A ∗ 02:01.
- Department of Biochemistry and Molecular Biology, Monash University, Clayton, VIC 3800, Australia.
Organizational Affiliation: