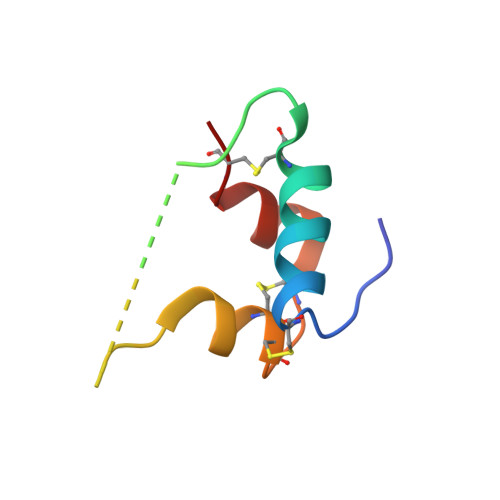
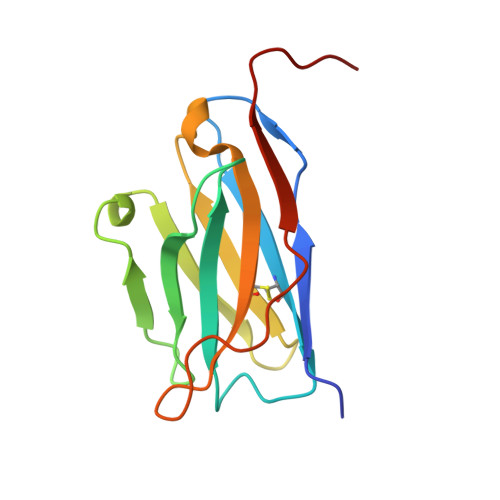
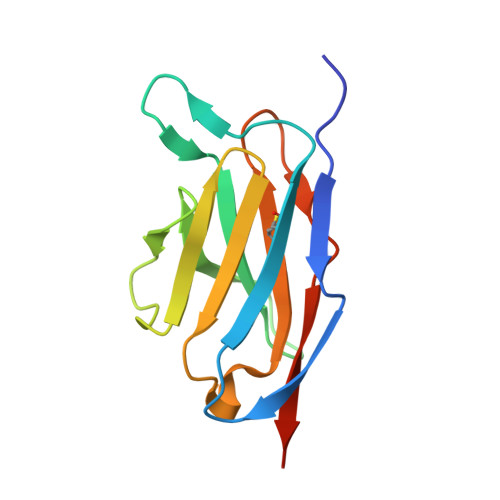
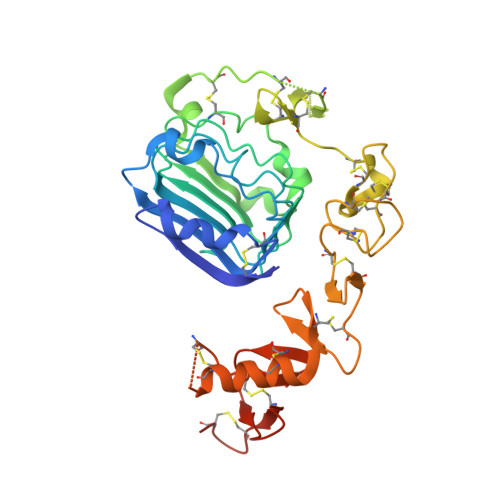
Single-chain insulin analogs threaded by the insulin receptor alpha CT domain.
Smith, N.A., Menting, J.G., Weiss, M.A., Lawrence, M.C., Smith, B.J.(2022) Biophys J
- PubMed: 36181268
- DOI: https://doi.org/10.1016/j.bpj.2022.09.038
- Primary Citation of Related Structures:
7KD6 - PubMed Abstract:
Insulin is a mainstay of therapy for diabetes mellitus, yet its thermal stability complicates global transportation and storage. Cold-chain transport, coupled with optimized formulation and materials, prevents to some degree nucleation of amyloid and hence inactivation of hormonal activity. These issues hence motivate the design of analogs with increased stability, with a promising approach being single-chain insulins (SCIs), whose C domains (foreshortened relative to proinsulin) resemble those of the single-chain growth factors (IGFs). We have previously demonstrated that optimized SCIs can exhibit native-like hormonal activity with enhanced thermal stability and marked resistance to fibrillation. Here, we describe the crystal structure of an ultrastable SCI (C-domain length 6; sequence EEGPRR) bound to modules of the insulin receptor (IR) ectodomain (N-terminal α-subunit domains L1-CR and C-terminal αCT peptide; "microreceptor" [μIR]). The structure of the SCI-μIR complex, stabilized by an Fv module, was determined using diffraction data to a resolution of 2.6 Å. Remarkably, the αCT peptide (IR-A isoform) "threads" through a gap between the flexible C domain and the insulin core. To explore such threading, we undertook molecular dynamics simulations to 1) compare threaded with unthreaded binding modes and 2) evaluate effects of C-domain length on these alternate modes. The simulations (employing both conventional and enhanced sampling simulations) provide evidence that very short linkers (C-domain length of -1) would limit gap opening in the SCI and so impair threading. We envisage that analogous threading occurs in the intact SCI-IR complex-rationalizing why minimal C-domain lengths block complete activity-and might be exploited to design novel receptor-isoform-specific analogs.
- La Trobe Institute for Molecular Science, La Trobe University, Melbourne, Victoria, Australia.
Organizational Affiliation: