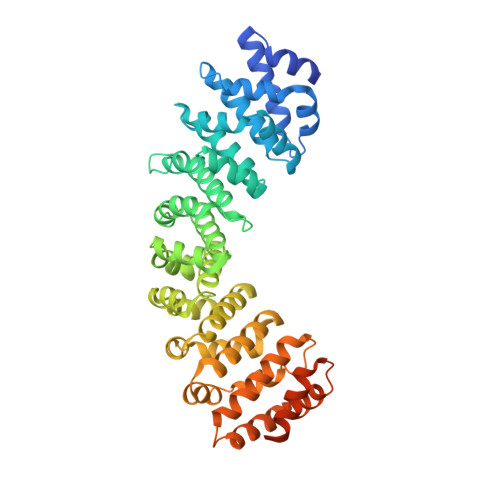
Targeting Nuclear LSD1 to Reprogram Cancer Cells and Reinvigorate Exhausted T Cells via a Novel LSD1-EOMES Switch.
Tu, W.J., McCuaig, R.D., Tan, A.H.Y., Hardy, K., Seddiki, N., Ali, S., Dahlstrom, J.E., Bean, E.G., Dunn, J., Forwood, J., Tsimbalyuk, S., Smith, K., Yip, D., Malik, L., Prasanna, T., Milburn, P., Rao, S.(2020) Front Immunol 11: 1228
- PubMed: 32612611
- DOI: https://doi.org/10.3389/fimmu.2020.01228
- Primary Citation of Related Structures:
7JJL - PubMed Abstract:
Lysine specific demethylase 1 (LSD1) is a key epigenetic eraser enzyme implicated in cancer metastases and recurrence. Nuclear LSD1 phosphorylated at serine 111 (nLSD1p) has been shown to be critical for the development of breast cancer stem cells. Here we show that circulating tumor cells isolated from immunotherapy-resistant metastatic melanoma patients express higher levels of nLSD1p compared to responders, which is associated with co-expression of stem-like, mesenchymal genes. Targeting nLSD1p with selective nLSD1 inhibitors better inhibits the stem-like mesenchymal signature than traditional FAD-specific LSD1 catalytic inhibitors such as GSK2879552. We also demonstrate that nLSD1p is enriched in PD-1 + CD8 + T cells from resistant melanoma patients and 4T1 immunotherapy-resistant mice. Targeting the LSD1p nuclear axis induces IFN-γ/TNF-α-expressing CD8 + T cell infiltration into the tumors of 4T1 immunotherapy-resistant mice, which is further augmented by combined immunotherapy. Underpinning these observations, nLSD1p is regulated by the key T cell exhaustion transcription factor EOMES in dysfunctional CD8 + T cells. EOMES co-exists with nLSD1p in PD-1 + CD8 + T cells in resistant patients, and nLSD1p regulates EOMES nuclear dynamics via demethylation/acetylation switching of critical EOMES residues. Using novel antibodies to target these post-translational modifications, we show that EOMES demethylation/acetylation is reciprocally expressed in resistant and responder patients. Overall, we show for the first time that dual inhibition of metastatic cancer cells and re-invigoration of the immune system requires LSD1 inhibitors that target the nLSD1p axis.
- Gene Regulation and Translational Medicine Laboratory, QIMR Berghofer Medical Research Institute, Brisbane, QLD, Australia.
Organizational Affiliation: