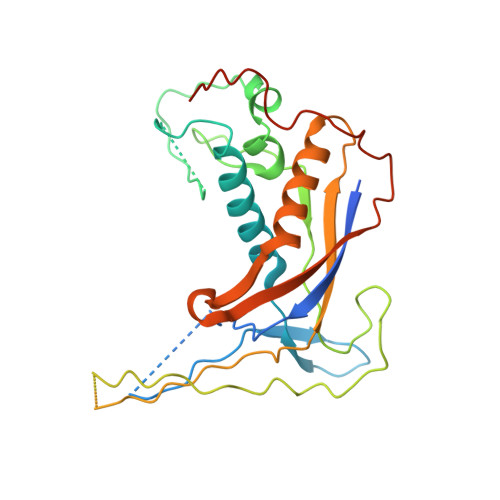
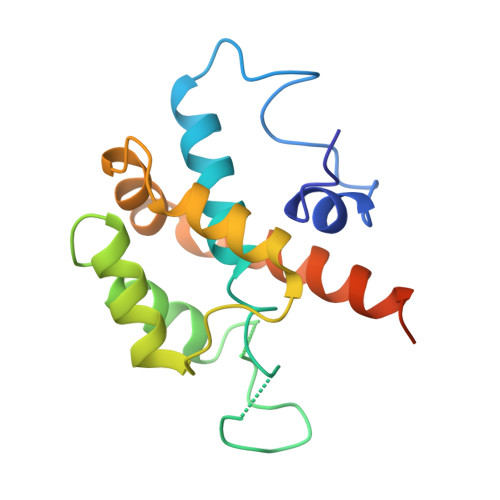
Structure of a type IV CRISPR-Cas ribonucleoprotein complex.
Zhou, Y., Bravo, J.P.K., Taylor, H.N., Steens, J.A., Jackson, R.N., Staals, R.H.J., Taylor, D.W.(2021) iScience 24: 102201-102201
- PubMed: 33733066
- DOI: https://doi.org/10.1016/j.isci.2021.102201
- Primary Citation of Related Structures:
7JHY - PubMed Abstract:
We reveal the cryo-electron microscopy structure of a type IV-B CRISPR ribonucleoprotein (RNP) complex (Csf) at 3.9-Å resolution. The complex best resembles the type III-A CRISPR Csm effector complex, consisting of a Cas7-like (Csf2) filament intertwined with a small subunit (Cas11) filament, but the complex lacks subunits for RNA processing and target DNA cleavage. Surprisingly, instead of assembling around a CRISPR-derived RNA (crRNA), the complex assembles upon heterogeneous RNA of a regular length arranged in a pseudo-A-form configuration. These findings provide a high-resolution glimpse into the assembly and function of enigmatic type IV CRISPR systems, expanding our understanding of class I CRISPR-Cas system architecture, and suggesting a function for type IV-B RNPs that may be distinct from other class 1 CRISPR-associated systems.
- Department of Molecular Biosciences, University of Texas at Austin, Austin, TX, USA.
Organizational Affiliation: