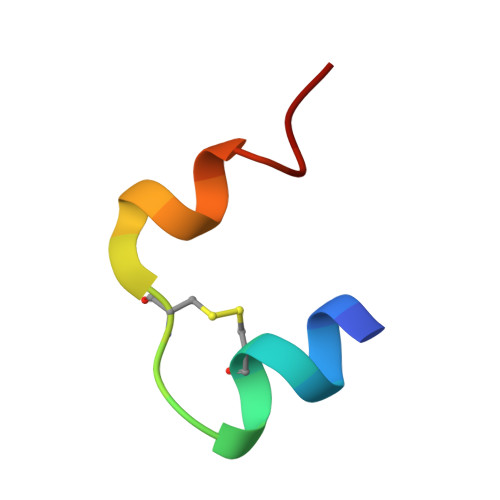
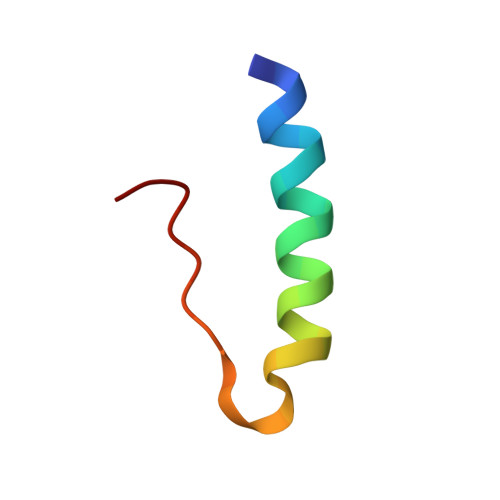
Structure of porcine insulin cocrystallized with clupeine Z.
Balschmidt, P., Hansen, F.B., Dodson, E.J., Dodson, G.G., Korber, F.(1991) Acta Crystallogr B 47: 975-986
- PubMed: 1772633
- DOI: https://doi.org/10.1107/s010876819100842x
- Primary Citation of Related Structures:
7INS - PubMed Abstract:
The crystal structure of NPH-insulin, pig insulin cocrystallized with zinc, m-cresol and protamine, has been solved by molecular replacement and refined using restrained least-squares refinement methods. The final crystallographic R factor for all reflections between 2 and 10 A is 19.4%. The insulin molecules are arranged as hexamers with two tetrahedrally coordinated Zn atoms in the central channel and one m-cresol bound to each monomer near His B5. One protamine binding site has been unequivocally identified near a dimer-dimer interface, although most of the polypeptide is crystallographically disordered. The conformation of the insulin moiety and the structural differences between the three unique monomers have been analysed. The zinc and m-cresol environments are described and the nature of the protamine binding site is outlined.
- Novo-Nordisk A/S, Gentofte, Denmark.
Organizational Affiliation: