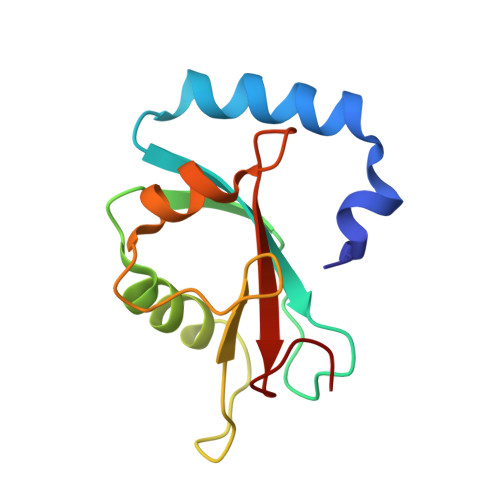
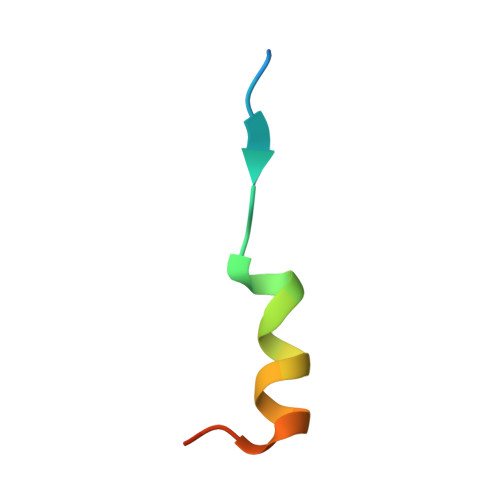
The crystal structure of the FAM134B-GABARAP complex provides mechanistic insights into the selective binding of FAM134 to the GABARAP subfamily.
Zhao, J., Li, Z., Li, J.(2022) FEBS Open Bio 12: 320-331
- PubMed: 34854256
- DOI: https://doi.org/10.1002/2211-5463.13340
- Primary Citation of Related Structures:
7FB5 - PubMed Abstract:
The mammalian Atg8 family (Atg8s proteins) consists of two subfamilies: GABARAP and LC3. All members can bind to the LC3-interacting region (LIR) or Atg8-interacting motif and participate in multiple steps of autophagy. The endoplasmic reticulum (ER) autophagy receptor FAM134B contains an LIR motif that can bind to Atg8s, but whether it can differentially bind to the two subfamilies and, if so, the structural basis for this preference remains unknown. Here, we found that FAM134B bound to the GABARAP subfamily more strongly than to the LC3 subfamily. We then solved the crystal structure of the FAM134B-GABARAP complex and demonstrated that FAM134B used both its LIR core and the C-terminal helix to bind to GABARAP. We further showed that these properties might be conserved in FAM134A or FAM134C. The structure also allowed us to identify the structural determinants for the binding selectivity. Our work may be valuable for studying the differential functions of GABARAP and LC3 subfamilies in ER phagy in future.
- Division of Cell, Developmental and Integrative Biology, School of Medicine, South China University of Technology, Guangzhou, China.
Organizational Affiliation: