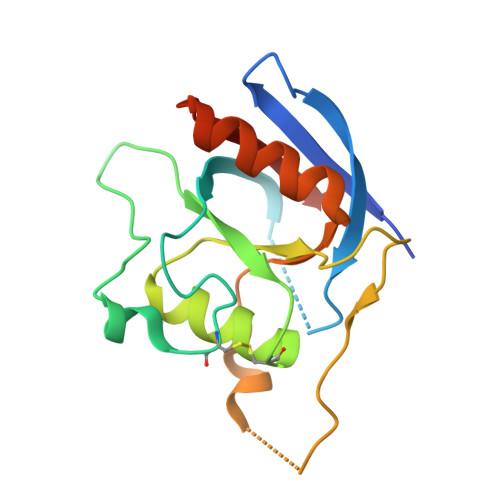
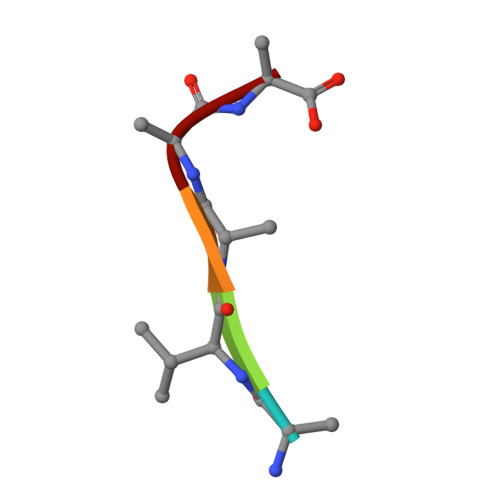
Structural insights into how vacuolar sorting receptors recognize the sorting determinants of seed storage proteins.
Tsao, H.E., Lui, S.N., Lo, A.H., Chen, S., Wong, H.Y., Wong, C.K., Jiang, L., Wong, K.B.(2022) Proc Natl Acad Sci U S A 119
- PubMed: 34983843
- DOI: https://doi.org/10.1073/pnas.2111281119
- Primary Citation of Related Structures:
7F2D, 7F2I - PubMed Abstract:
In Arabidopsis , vacuolar sorting receptor isoform 1 (VSR1) sorts 12S globulins to the protein storage vacuoles during seed development. Vacuolar sorting is mediated by specific protein-protein interactions between VSR1 and the vacuolar sorting determinant located at the C terminus (ctVSD) on the cargo proteins. Here, we determined the crystal structure of the protease-associated domain of VSR1 (VSR1-PA) in complex with the C-terminal pentapeptide ( 468 RVAAA 472 ) of cruciferin 1, an isoform of 12S globulins. The 468 RVA 470 motif forms a parallel β-sheet with the switch III residues ( 127 TMD 129 ) of VSR1-PA, and the 471 AA 472 motif docks to a cradle formed by the cargo-binding loop ( 95 RGDCYF 100 ), making a hydrophobic interaction with Tyr99. The C-terminal carboxyl group of the ctVSD is recognized by forming salt bridges with Arg95. The C-terminal sequences of cruciferin 1 and vicilin-like storage protein 22 were sufficient to redirect the secretory red fluorescent protein (spRFP) to the vacuoles in Arabidopsis protoplasts. Adding a proline residue to the C terminus of the ctVSD and R95M substitution of VSR1 disrupted receptor-cargo interactions in vitro and led to increased secretion of spRFP in Arabidopsis protoplasts. How VSR1-PA recognizes ctVSDs of other storage proteins was modeled. The last three residues of ctVSD prefer hydrophobic residues because they form a hydrophobic cluster with Tyr99 of VSR1-PA. Due to charge-charge interactions, conserved acidic residues, Asp129 and Glu132, around the cargo-binding site should prefer basic residues over acidic ones in the ctVSD. The structural insights gained may be useful in targeting recombinant proteins to the protein storage vacuoles in seeds.
- Centre for Protein Science and Crystallography, School of Life Sciences, The Chinese University of Hong Kong (CUHK), Shatin, Hong Kong, China.
Organizational Affiliation: