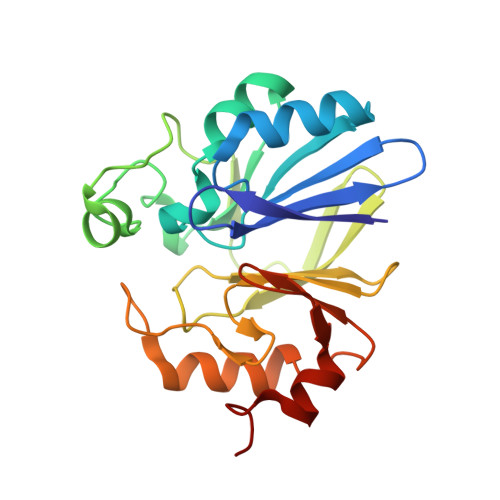
Dual Activity BLEG-1 from Bacillus lehensis G1 Revealed Structural Resemblance to B3 Metallo-beta-Lactamase and Glyoxalase II: An Insight into Its Enzyme Promiscuity and Evolutionary Divergence.
Au, S.X., Dzulkifly, N.S., Muhd Noor, N.D., Matsumura, H., Raja Abdul Rahman, R.N.Z., Normi, Y.M.(2021) Int J Mol Sci 22
- PubMed: 34502284
- DOI: https://doi.org/10.3390/ijms22179377
- Primary Citation of Related Structures:
7EV5 - PubMed Abstract:
Metallo-β-lactamases (MBLs) are class B β-lactamases from the metallo-hydrolase-like MBL-fold superfamily which act on a broad range of β-lactam antibiotics. A previous study on BLEG-1 (formerly called Bleg1_2437), a hypothetical protein from Bacillus lehensis G1, revealed sequence similarity and activity to B3 subclass MBLs, despite its evolutionary divergence from these enzymes. Its relatedness to glyoxalase II (GLXII) raises the possibility of its enzymatic promiscuity and unique structural features compared to other MBLs and GLXIIs. This present study highlights that BLEG-1 possessed both MBL and GLXII activities with similar catalytic efficiencies. Its crystal structure revealed highly similar active site configuration to YcbL and GloB GLXIIs from Salmonella enterica , and L1 B3 MBL from Stenotrophomonas maltophilia . However, different from GLXIIs, BLEG-1 has an insertion of an active-site loop, forming a binding cavity similar to B3 MBL at the N-terminal region. We propose that BLEG-1 could possibly have evolved from GLXII and adopted MBL activity through this insertion.
- Enzyme and Microbial Technology (EMTech) Research Center, Faculty of Biotechnology and Biomolecular Sciences, Universiti Putra Malaysia, Serdang 43400, Selangor, Malaysia.
Organizational Affiliation: