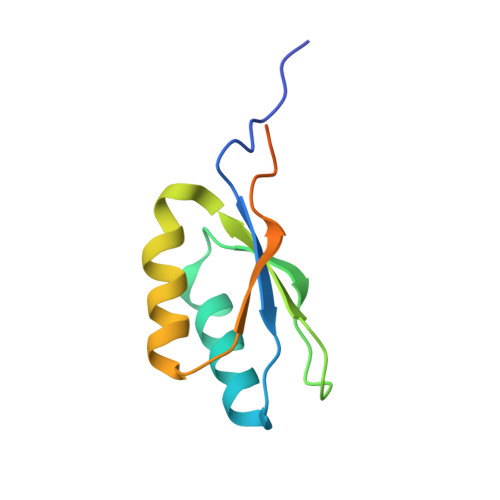
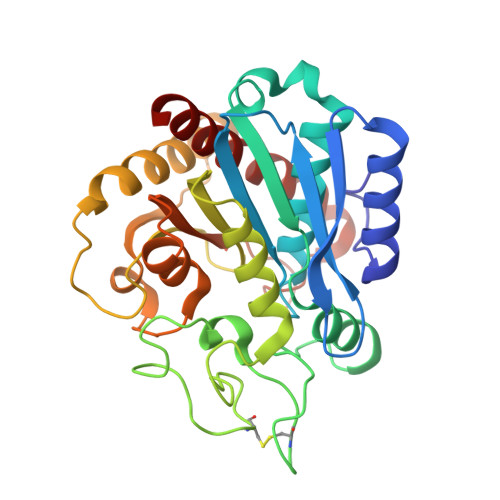
Structure of Aedes aegypti procarboxypeptidase B1 and its binding with Dengue virus for controlling infection.
Gavor, E., Choong, Y.K., Tulsian, N.K., Nayak, D., Idris, F., Sivaraman, H., Ting, D.H.R., Sylvie, A., Mok, Y.K., Kini, R.M., Sivaraman, J.(2022) Life Sci Alliance 5
- PubMed: 34750241
- DOI: https://doi.org/10.26508/lsa.202101211
- Primary Citation of Related Structures:
7EQX - PubMed Abstract:
Metallocarboxypeptidases play critical roles in the development of mosquitoes and influence pathogen/parasite infection of the mosquito midgut. Here, we report the crystal structure of Aedes aegypti procarboxypeptidase B1 (PCPBAe1), characterized its substrate specificity and mechanism of binding to and inhibiting Dengue virus (DENV). We show that the activated PCPBAe1 (CPBAe1) hydrolyzes both Arg- and Lys-substrates, which is modulated by residues Asp 251 and Ser 239 Notably, these residues are conserved in CPBs across mosquito species, possibly required for efficient digestion of basic dietary residues that are necessary for mosquito reproduction and development. Importantly, we characterized the interaction between PCPBAe1 and DENV envelope (E) protein, virus-like particles, and infectious virions. We identified residues Asp 18A , Glu 19A , Glu 85 , Arg 87 , and Arg 89 of PCPBAe1 are essential for interaction with DENV. PCPBAe1 maps to the dimeric interface of the E protein domains I/II (Lys 64 -Glu 84 , Val 238 -Val 252 , and Leu 278 -Leu 287 ). Overall, our studies provide general insights into how the substrate-binding property of mosquito carboxypeptidases could be targeted to potentially control mosquito populations or proposes a mechanism by which PCPBAe1 binds to and inhibits DENV.
- Department of Biological Sciences, National University of Singapore, Singapore.
Organizational Affiliation: