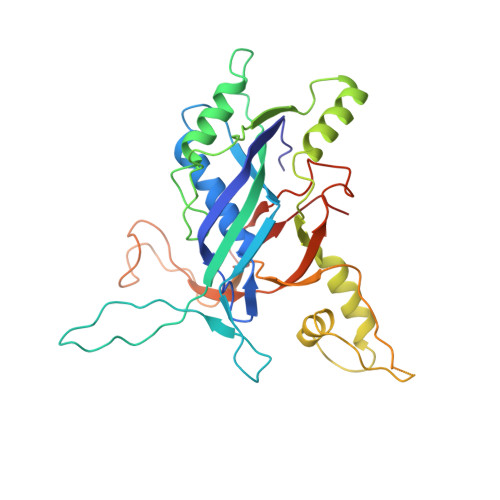
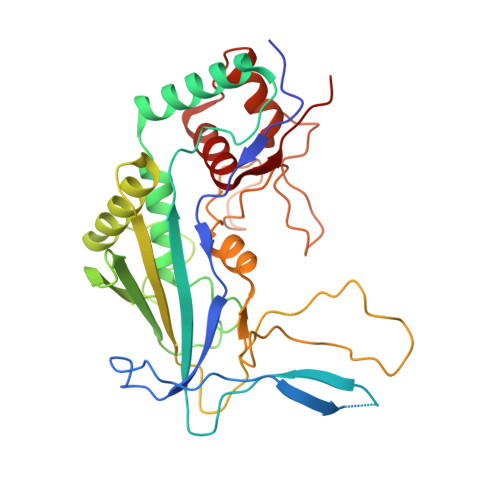
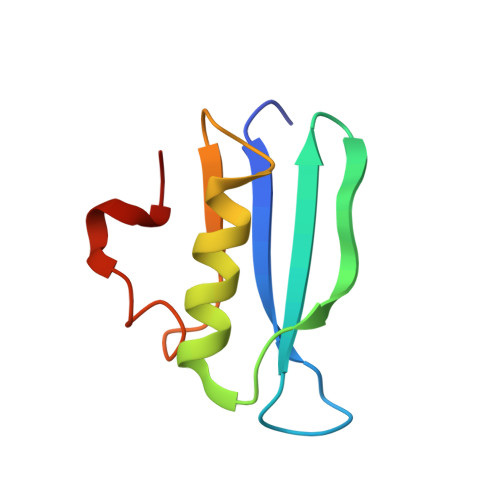
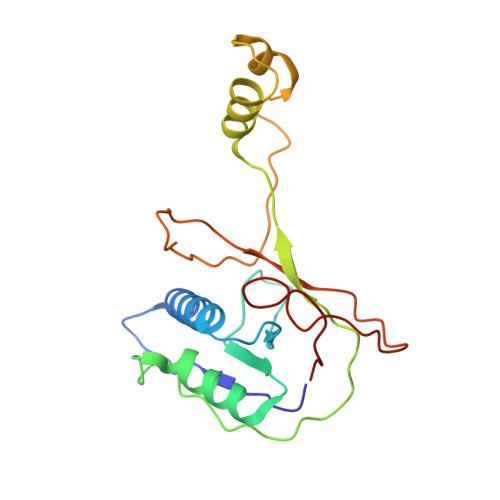
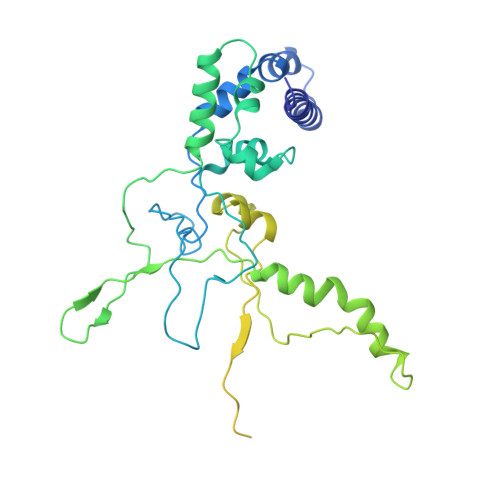
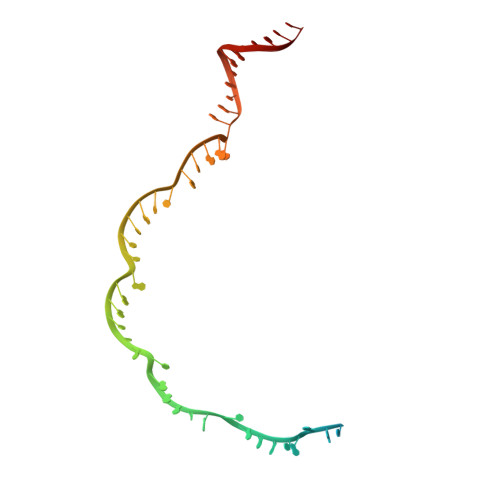
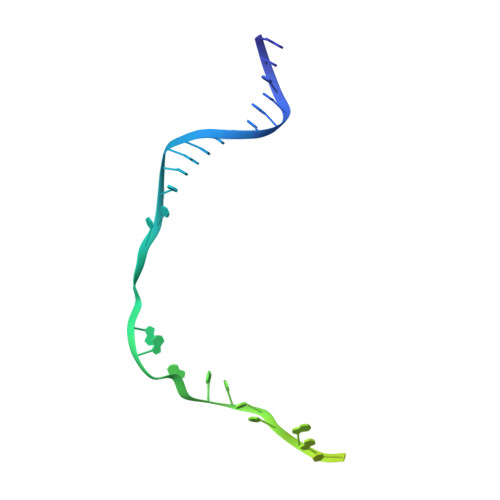
AcrIF5 specifically targets DNA-bound CRISPR-Cas surveillance complex for inhibition.
Xie, Y., Zhang, L., Gao, Z., Yin, P., Wang, H., Li, H., Chen, Z., Zhang, Y., Yang, M., Feng, Y.(2022) Nat Chem Biol 18: 670-677
- PubMed: 35301482
- DOI: https://doi.org/10.1038/s41589-022-00995-8
- Primary Citation of Related Structures:
7EQG, 7F45 - PubMed Abstract:
CRISPR-Cas systems are prokaryotic antiviral systems, and phages use anti-CRISPR proteins (Acrs) to inactivate these systems. Here we present structural and functional analyses of AcrIF5, exploring its unique anti-CRISPR mechanism. AcrIF5 shows binding specificity only for the target DNA-bound form of the crRNA-guided surveillance (Csy) complex, but not the apo Csy complex from the type I-F CRISPR-Cas system. We solved the structure of the Csy-dsDNA-AcrIF5 complex, revealing that the conformational changes of the Csy complex caused by dsDNA binding dictate the binding specificity for the Csy-dsDNA complex by AcrIF5. Mechanistically, five AcrIF5 molecules bind one Csy-dsDNA complex, which destabilizes the helical bundle domain of Cas8f, thus preventing subsequent Cas2/3 recruitment. AcrIF5 exists in symbiosis with AcrIF3, which blocks Cas2/3 recruitment. This attack on the recruitment event stands in contrast to the conventional mechanisms of blocking binding of target DNA. Overall, our study reveals an unprecedented mechanism of CRISPR-Cas inhibition by AcrIF5.
- Beijing Advanced Innovation Center for Soft Matter Science and Engineering, Beijing Key Laboratory of Bioprocess, State Key Laboratory of Chemical Resource Engineering, College of Life Science and Technology, Beijing University of Chemical Technology, Beijing, China.
Organizational Affiliation: