Crystal Structure of Signal Recognition Particle 54 kDa protein (SRP54) from Aeropyrum pernix K1 in Complex with GDP
Xie, Y., Zhang, B., Liu, J.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Signal recognition particle 54 kDa protein | 441 | Aeropyrum pernix K1 | Mutation(s): 0 Gene Names: srp54, APE_1735 EC: 3.6.5.4 | 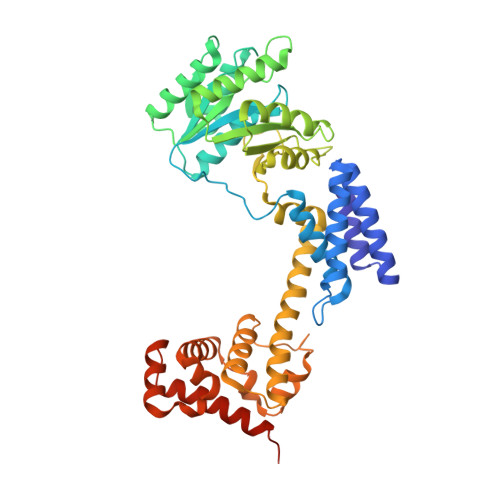 | |
UniProt | |||||
Find proteins for Q9YB62 (Aeropyrum pernix (strain ATCC 700893 / DSM 11879 / JCM 9820 / NBRC 100138 / K1)) Explore Q9YB62 Go to UniProtKB: Q9YB62 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9YB62 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| GDP (Subject of Investigation/LOI) Query on GDP | B [auth A] | GUANOSINE-5'-DIPHOSPHATE C10 H15 N5 O11 P2 QGWNDRXFNXRZMB-UUOKFMHZSA-N |  | ||
| MG (Subject of Investigation/LOI) Query on MG | C [auth A] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 82.498 | α = 90 |
| b = 82.498 | β = 90 |
| c = 140.805 | γ = 120 |
| Software Name | Purpose |
|---|---|
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| REFMAC | refinement |
| PDB_EXTRACT | data extraction |
| BUCCANEER | phasing |