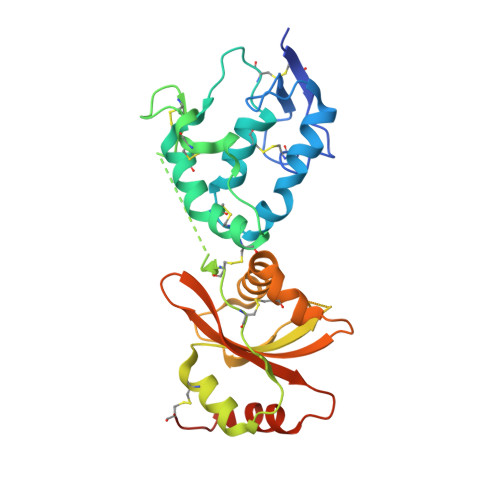
Dynamics of the secreted frizzled related protein Sizzled and potential implications for binding to bone morphogenetic protein-1 (BMP-1).
Sharma, U., Vadon-Le Goff, S., Harlos, K., Zhao, Y., Mariano, N., Bijakowski, C., Bourhis, J.M., Moali, C., Hulmes, D.J.S., Aghajari, N.(2022) Sci Rep 12: 14850-14850
- PubMed: 36050373
- DOI: https://doi.org/10.1038/s41598-022-18795-4
- Primary Citation of Related Structures:
7EL5 - PubMed Abstract:
Sizzled (Szl) is both a secreted frizzled related protein (sFRP) and a naturally occurring inhibitor of the zinc metalloproteinase bone morphogenetic protein-1 (BMP-1), a key regulator of extracellular matrix assembly and growth factor activation. Here we present a new crystal structure for Szl which differs from that previously reported by a large scale (90°) hinge rotation between its cysteine-rich and netrin-like domains. We also present results of a molecular docking analysis showing interactions likely to be involved in the inhibition of BMP-1 activity by Szl. When compared with known structures of BMP-1 in complex with small molecule inhibitors, this reveals features that may be helpful in the design of new inhibitors to prevent the excessive accumulation of extracellular matrix that is the hallmark of fibrotic diseases.
- Molecular Microbiology and Structural Biochemistry, UMR 5086 CNRS-University of Lyon, 7 passage du Vercors, 69367, Lyon, France.
Organizational Affiliation: