Peptide Presentations of Marsupial MHC Class I Visualize Immune Features of Lower Mammals Paralleled with Bats.
Wang, P., Yue, C., Liu, K., Lu, D., Liu, S., Yao, S., Li, X., Su, X., Ren, K., Chai, Y., Qi, J., Zhao, Y., Lou, Y., Sun, Z., Gao, G.F., Liu, W.J.(2021) J Immunol 207: 2167-2178
- PubMed: 34535575
- DOI: https://doi.org/10.4049/jimmunol.2100350
- Primary Citation of Related Structures:
7EDO - PubMed Abstract:
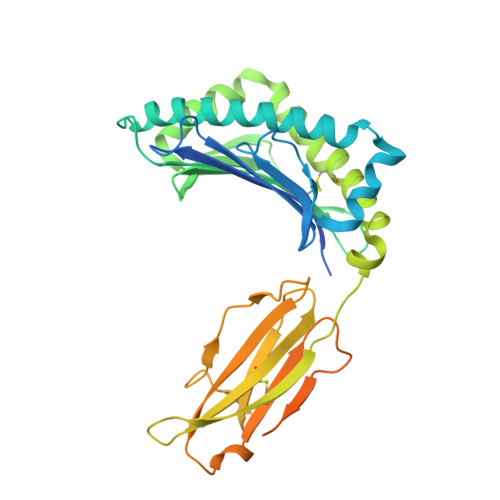
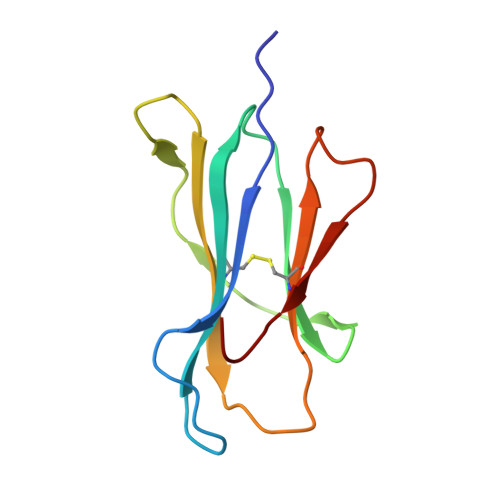
Marsupials are one of three major mammalian lineages that include the placental eutherians and the egg-laying monotremes. The marsupial brushtail possum is an important protected species in the Australian forest ecosystem. Molecules encoded by the MHC genes are essential mediators of adaptive immune responses in virus-host interactions. Yet, nothing is known about the peptide presentation features of any marsupial MHC class I (MHC I). This study identified a series of possum MHC I Trvu-UB*01:01 binding peptides derived from wobbly possum disease virus (WPDV), a lethal virus of both captive and feral possum populations, and unveiled the structure of marsupial peptide/MHC I complex. Notably, we found the two brushtail possum-specific insertions, the 3-aa Ile 52 Glu 53 Arg 54 and 1-aa Arg 154 insertions are located in the Trvu-UB*01:01 peptide binding groove (PBG). The 3-aa insertion plays a pivotal role in maintaining the stability of the N terminus of Trvu-UB*01:01 PBG. This aspect of marsupial PBG is unexpectedly similar to the bat MHC I Ptal-N*01:01 and is shared with lower vertebrates from elasmobranch to monotreme, indicating an evolution hotspot that may have emerged from the pathogen-host interactions. Residue Arg 154 insertion, located in the α2 helix, is available for TCR recognition, and it has a particular influence on promoting the anchoring of peptide WPDV-12. These findings add significantly to our understanding of adaptive immunity in marsupials and its evolution in vertebrates. Our findings have the potential to impact the conservation of the protected species brushtail possum and other marsupial species.
- School of Laboratory Medicine and Life Sciences, Wenzhou Medical University, Wenzhou, China.
Organizational Affiliation: