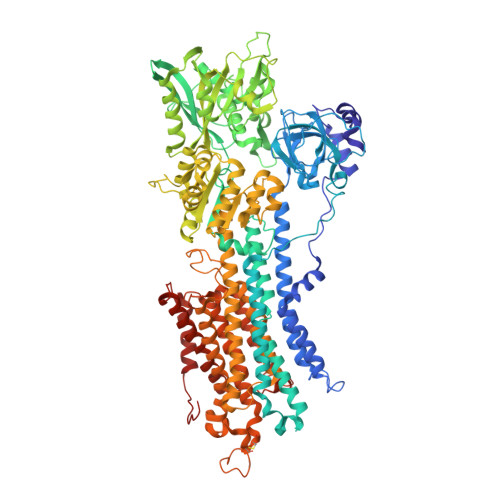
Cryo-EM analysis provides new mechanistic insight into ATP binding to Ca 2+ -ATPase SERCA2b.
Zhang, Y., Watanabe, S., Tsutsumi, A., Kadokura, H., Kikkawa, M., Inaba, K.(2021) EMBO J 40: e108482-e108482
- PubMed: 34459010
- DOI: https://doi.org/10.15252/embj.2021108482
- Primary Citation of Related Structures:
7E7S - PubMed Abstract:
Sarco/endoplasmic reticulum Ca 2+ -ATPase (SERCA) 2b is a ubiquitous SERCA family member that conducts Ca 2+ uptake from the cytosol to the ER. Herein, we present a 3.3 Å resolution cryo-electron microscopy (cryo-EM) structure of human SERCA2b in the E1·2Ca 2+ state, revealing a new conformation for Ca 2+ -bound SERCA2b with a much closer arrangement of cytosolic domains than in the previously reported crystal structure of Ca 2+ -bound SERCA1a. Multiple conformations generated by 3D classification of cryo-EM maps reflect the intrinsically dynamic nature of the cytosolic domains in this state. Notably, ATP binding residues of SERCA2b in the E1·2Ca 2+ state are located at similar positions to those in the E1·2Ca 2+ -ATP state; hence, the cryo-EM structure likely represents a preformed state immediately prior to ATP binding. Consistently, a SERCA2b mutant with an interdomain disulfide bridge that locks the closed cytosolic domain arrangement displayed significant autophosphorylation activity in the presence of Ca 2+ . We propose a novel mechanism of ATP binding to SERCA2b.
- Institute of Multidisciplinary Research for Advanced Materials, Tohoku University, Sendai, Japan.
Organizational Affiliation: