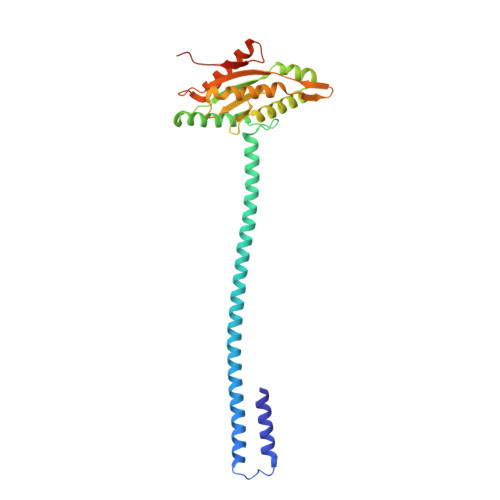
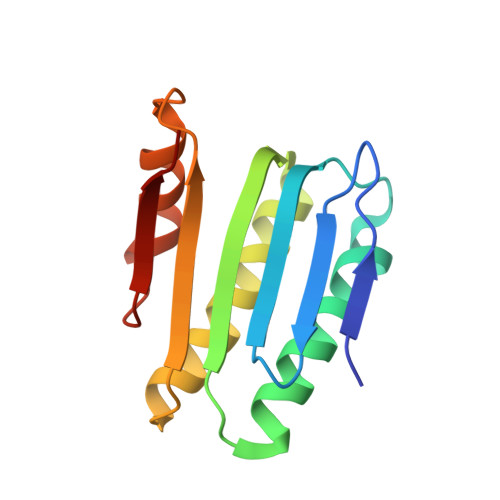
Structural basis for diguanylate cyclase activation by its binding partner in Pseudomonas aeruginosa .
Chen, G., Zhou, J., Zuo, Y., Huo, W., Peng, J., Li, M., Zhang, Y., Wang, T., Zhang, L., Zhang, L., Liang, H.(2021) Elife 10
- PubMed: 34498587
- DOI: https://doi.org/10.7554/eLife.67289
- Primary Citation of Related Structures:
7E6G - PubMed Abstract:
Cyclic-di-guanosine monophosphate (c-di-GMP) is an important effector associated with acute-chronic infection transition in Pseudomonas aeruginosa . Previously, we reported a signaling network SiaABCD, which regulates biofilm formation by modulating c-di-GMP level. However, the mechanism for SiaD activation by SiaC remains elusive. Here we determine the crystal structure of SiaC-SiaD-GpCpp complex and revealed a unique mirror symmetric conformation: two SiaD form a dimer with long stalk domains, while four SiaC bind to the conserved motifs on the stalks of SiaD and stabilize the conformation for further enzymatic catalysis. Furthermore, SiaD alone exhibits an inactive pentamer conformation in solution, demonstrating that SiaC activates SiaD through a dynamic mechanism of promoting the formation of active SiaD dimers. Mutagenesis assay confirmed that the stalks of SiaD are necessary for its activation. Together, we reveal a novel mechanism for DGC activation, which clarifies the regulatory networks of c-di-GMP signaling.
- Key Laboratory of Resources Biology and Biotechnology in Western China, Ministry of Education, College of Life Sciences, Northwest University, ShaanXi, China.
Organizational Affiliation: