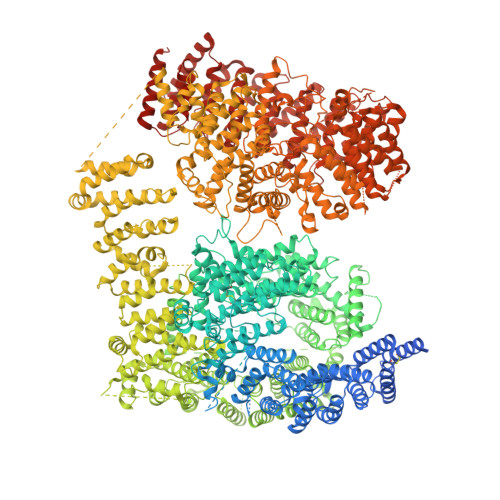
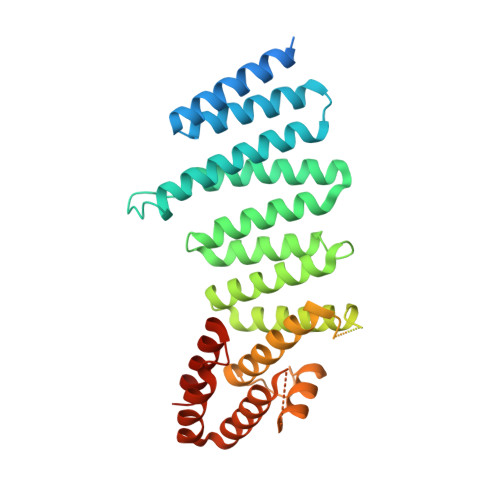
Pathological polyQ expansion does not alter the conformation of the Huntingtin-HAP40 complex.
Huang, B., Guo, Q., Niedermeier, M.L., Cheng, J., Engler, T., Maurer, M., Pautsch, A., Baumeister, W., Stengel, F., Kochanek, S., Fernandez-Busnadiego, R.(2021) Structure 29: 804-809.e5
- PubMed: 33909994
- DOI: https://doi.org/10.1016/j.str.2021.04.003
- Primary Citation of Related Structures:
7DXJ, 7DXK - PubMed Abstract:
The abnormal amplification of a CAG repeat in the gene coding for huntingtin (HTT) leads to Huntington's disease (HD). At the protein level, this translates into the expansion of a polyglutamine (polyQ) stretch located at the HTT N terminus, which renders HTT aggregation prone by unknown mechanisms. Here we investigated the effects of polyQ expansion on HTT in a complex with its stabilizing interaction partner huntingtin-associated protein 40 (HAP40). Surprisingly, our comprehensive biophysical, crosslinking mass spectrometry and cryo-EM experiments revealed no major differences in the conformation of HTT-HAP40 complexes of various polyQ length, including 17QHTT-HAP40 (wild type), 46QHTT-HAP40 (typical polyQ length in HD patients), and 128QHTT-HAP40 (extreme polyQ length). Thus, HTT polyQ expansion does not alter the global conformation of HTT when associated with HAP40.
- Department of Gene Therapy, Ulm University, 89081, Ulm, Germany.
Organizational Affiliation: