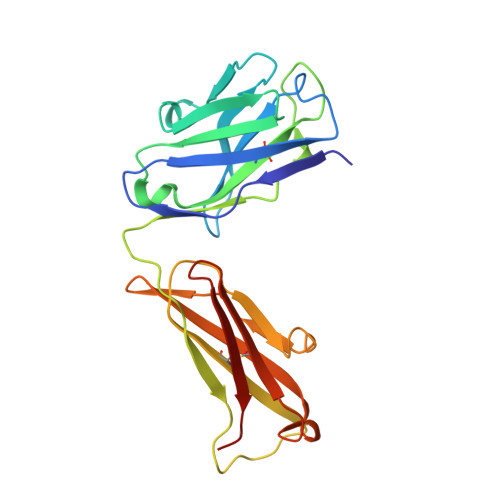
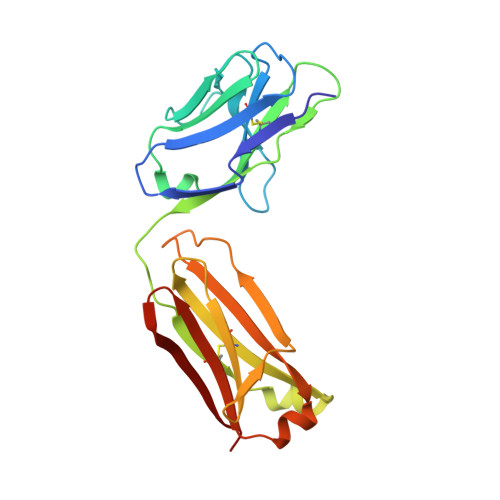
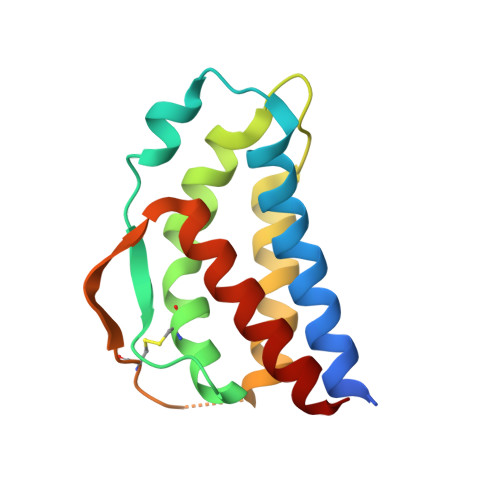
Crystal structure of human interleukin-2 in complex with TCB2, a new antibody-drug candidate with antitumor activity.
Kim, J., Lee, J.Y., Park, S.Y., Lee, Y.J., Kim, M.S.(2021) Oncoimmunology 10: 1899671-1899671
- PubMed: 33796411
- DOI: https://doi.org/10.1080/2162402X.2021.1899671
- Primary Citation of Related Structures:
7DR4 - PubMed Abstract:
Immunotherapy via interleukin-2 (IL-2) mediated activation of anti-tumor immune response is a promising approach for cancer treatment. The multi-potent cytokine, IL-2 has a central role in immune cell activation and homeostasis. Since IL-2 preferentially activates immunosuppressive T regulatory cells by IL-2Rα dependent manner, blocking IL-2:IL-2Rα interaction is a key to amplify the IL-2 activity in effector T cells toward anti-tumor response. Anti-IL-2 monoclonal antibodies are good candidates to control the IL-2:IL-2Rα interaction. In a previous study, we developed a new IL-2Rα mimetic antibody, TCB2, and showed that the human IL-2(hIL-2):TCB2 complex can stimulate T effector cells specifically and elicit potent anti-cancer immunotherapeutic effect, especially when administered in combination with immune checkpoint inhibitors. To understand the molecular mechanism, we determined the crystal structure of TCB2-Fab in a complex with hIL-2 at 2.5 Å resolution. Our structural analysis reveals that TCB2 binds to the central area of the hIL-2Rα binding region on hIL-2, and binding angle and epitope are different from previously known hIL-2Rα mimicking antibody NARA1 which recognizes the top part of hIL-2. TCB2 binding to hIL-2 also induces an allosteric effect that increases the affinity for the hetero-dimeric hIL-2 receptor, IL-2R(β + γ), on effector T cells.
- Department of Life Sciences, Pohang University of Science and Technology (POSTECH), Pohang, Gyeongbuk, Republic of Korea.
Organizational Affiliation: