Human antibodies targeting a Mycobacterium transporter protein mediate protection against tuberculosis.
Watson, A., Li, H., Ma, B., Weiss, R., Bendayan, D., Abramovitz, L., Ben-Shalom, N., Mor, M., Pinko, E., Bar Oz, M., Wang, Z., Du, F., Lu, Y., Rybniker, J., Dahan, R., Huang, H., Barkan, D., Xiang, Y., Javid, B., Freund, N.T.(2021) Nat Commun 12: 602-602
- PubMed: 33504803
- DOI: https://doi.org/10.1038/s41467-021-20930-0
- Primary Citation of Related Structures:
7DM1, 7DM2 - PubMed Abstract:
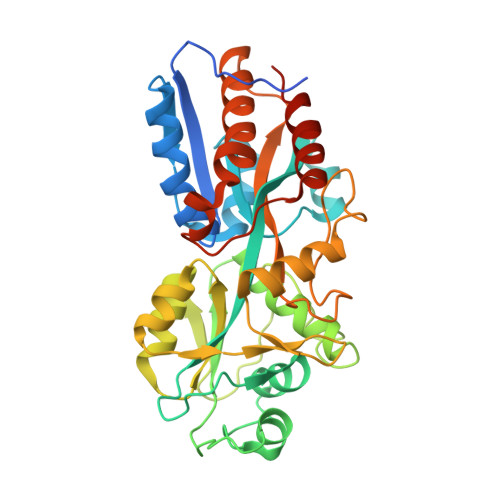
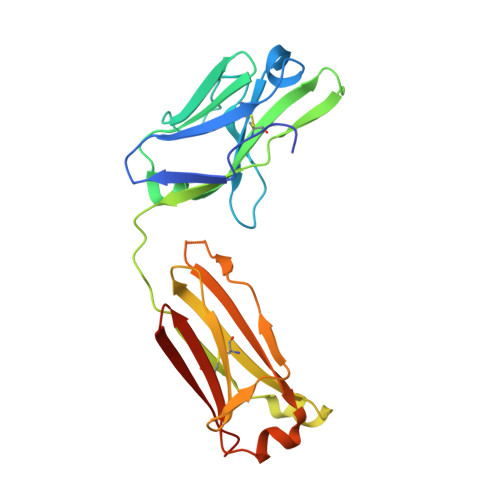
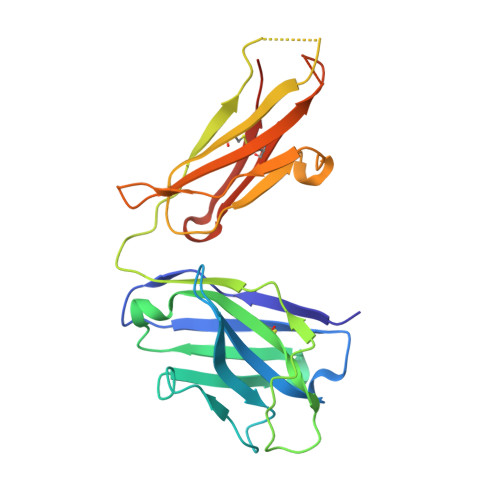
Mycobacterium tuberculosis (Mtb) exposure drives antibody responses, but whether patients with active tuberculosis elicit protective antibodies, and against which antigens, is still unclear. Here we generate monoclonal antibodies from memory B cells of one patient to investigate the B cell responses during active infection. The antibodies, members of four distinct B cell clones, are directed against the Mtb phosphate transporter subunit PstS1. Antibodies p4-36 and p4-163 reduce Mycobacterium bovis-BCG and Mtb levels in an ex vivo human whole blood growth inhibition assay in an FcR-dependent manner; meanwhile, germline versions of p4-36 and p4-163 do not bind Mtb. Crystal structures of p4-36 and p4-170, complexed to PstS1, are determined at 2.1 Å and 2.4 Å resolution, respectively, to reveal two distinctive PstS1 epitopes. Lastly, a prophylactic p4-36 and p4-163 treatment in Mtb-infected Balb/c mice reduces bacterial lung burden by 50%. Our study shows that inhibitory anti-PstS1 B cell responses arise during active tuberculosis.
- Department of Clinical Microbiology and Immunology, Sackler Faculty of Medicine, Tel Aviv University, Tel Aviv-Yafo, Israel.
Organizational Affiliation: