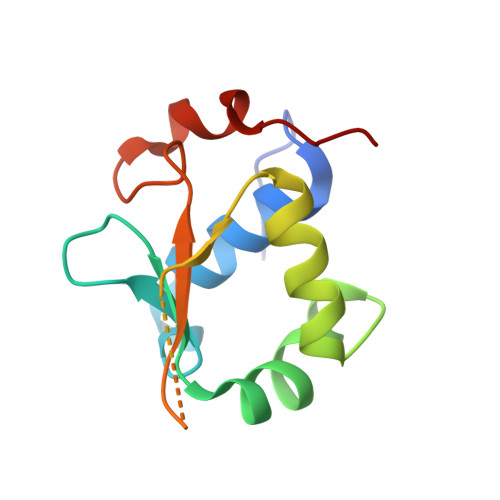
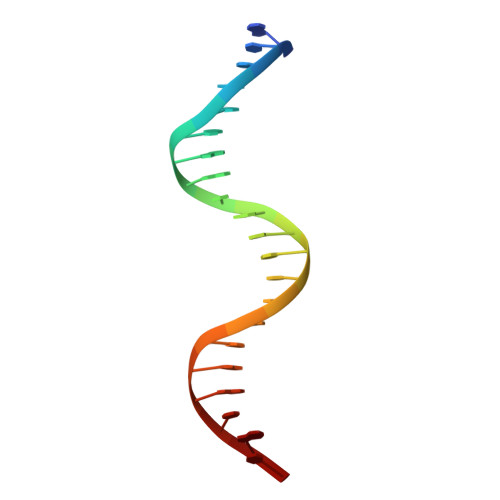
Structures of heat shock factor trimers bound to DNA.
Feng, N., Feng, H., Wang, S., Punekar, A.S., Ladenstein, R., Wang, D.C., Zhang, Q., Ding, J., Liu, W.(2021) iScience 24: 102951-102951
- PubMed: 34458700
- DOI: https://doi.org/10.1016/j.isci.2021.102951
- Primary Citation of Related Structures:
7DCJ, 7DCS, 7DCT, 7DCU - PubMed Abstract:
Heat shock factor 1 (HSF1) and 2 (HSF2) play distinct but overlapping regulatory roles in maintaining cellular proteostasis or mediating cell differentiation and development. Upon activation, both HSFs trimerize and bind to heat shock elements (HSEs) present in the promoter region of target genes. Despite structural insights gained from recent studies, structures reflecting the physiological architecture of this transcriptional machinery remains to be determined. Here, we present co-crystal structures of human HSF1 and HSF2 trimers bound to DNA, which reveal a triangular arrangement of the three DNA-binding domains (DBDs) with protein-protein interactions largely mediated by the wing domain. Two structural properties, different flexibility of the wing domain and local DNA conformational changes induced by HSF binding, seem likely to contribute to the subtle differential specificity between HSF1 and HSF2. Besides, two more structures showing DBDs bound to "two-site" head-to-head HSEs were determined as additions to the published tail-to-tail dimer-binding structures.
- National Laboratory of Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China.
Organizational Affiliation: