Mechanistic insights into the regulation of plant phosphate homeostasis by the rice SPX2 - PHR2 complex.
Guan, Z., Zhang, Q., Zhang, Z., Zuo, J., Chen, J., Liu, R., Savarin, J., Broger, L., Cheng, P., Wang, Q., Pei, K., Zhang, D., Zou, T., Yan, J., Yin, P., Hothorn, M., Liu, Z.(2022) Nat Commun 13: 1581-1581
- PubMed: 35332155
- DOI: https://doi.org/10.1038/s41467-022-29275-8
- Primary Citation of Related Structures:
7D3T, 7D3Y - PubMed Abstract:
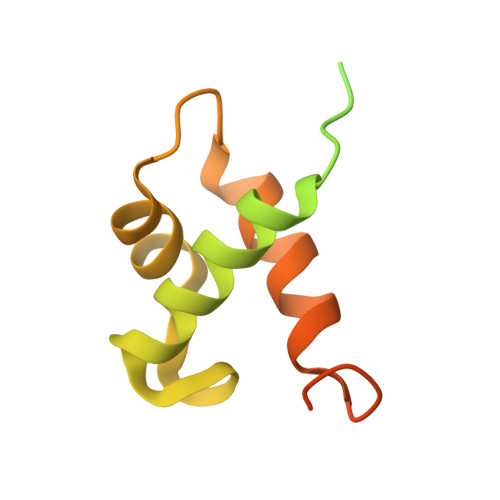
Phosphate (Pi) starvation response (PHR) transcription factors play key roles in plant Pi homeostasis maintenance. They are negatively regulated by stand-alone SPX proteins, cellular receptors for inositol pyrophosphate (PP-InsP) nutrient messengers. How PP-InsP-bound SPX interacts with PHRs is poorly understood. Here, we report crystal structures of the rice SPX2/InsP 6 /PHR2 complex and of the PHR2 DNA binding (MYB) domain in complex with target DNA at resolutions of 3.1 Å and 2.7 Å, respectively. In the SPX2/InsP 6 /PHR2 complex, the signalling-active SPX2 assembles into a domain-swapped dimer conformation and binds two copies of PHR2, targeting both its coiled-coil (CC) oligomerisation domain and MYB domain. Our results reveal that the SPX2 senses PP-InsPs to inactivate PHR2 by establishing severe steric clashes with the PHR2 MYB domain, preventing DNA binding, and by disrupting oligomerisation of the PHR2 CC domain, attenuating promoter binding. Our findings rationalize how PP-InsPs activate SPX receptor proteins to target PHR family transcription factors.
- National Key Laboratory of Crop Genetic Improvement, Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan, 430070, China.
Organizational Affiliation: