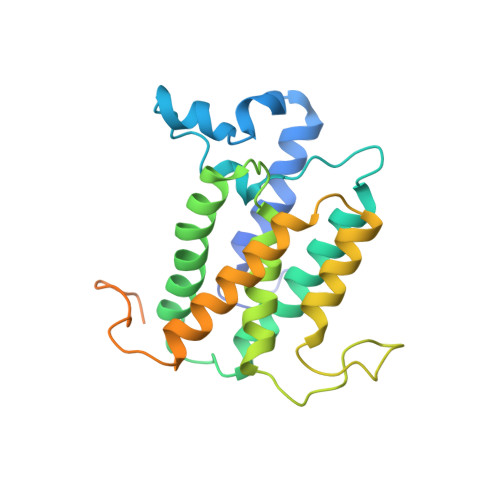
The cryo-EM structure of an ERAD protein channel formed by tetrameric human Derlin-1.
Rao, B., Li, S., Yao, D., Wang, Q., Xia, Y., Jia, Y., Shen, Y., Cao, Y.(2021) Sci Adv 7
- PubMed: 33658201
- DOI: https://doi.org/10.1126/sciadv.abe8591
- Primary Citation of Related Structures:
7CZB - PubMed Abstract:
Endoplasmic reticulum-associated degradation (ERAD) is a process directing misfolded proteins from the ER lumen and membrane to the degradation machinery in the cytosol. A key step in ERAD is the translocation of ER proteins to the cytosol. Derlins are essential for protein translocation in ERAD, but the mechanism remains unclear. Here, we solved the structure of human Derlin-1 by cryo-electron microscopy. The structure shows that Derlin-1 forms a homotetramer that encircles a large tunnel traversing the ER membrane. The tunnel has a diameter of about 12 to 15 angstroms, large enough to allow an α helix to pass through. The structure also shows a lateral gate within the membrane, providing access of transmembrane proteins to the tunnel, and thus, human Derlin-1 forms a protein channel for translocation of misfolded proteins. Our structure is different from the monomeric yeast Derlin structure previously reported, which forms a semichannel with another protein.
- CAS Center for Excellence in Molecular Cell Science, Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, University of Chinese Academy of Sciences, 333 Haike Road, Shanghai 201210, China.
Organizational Affiliation: