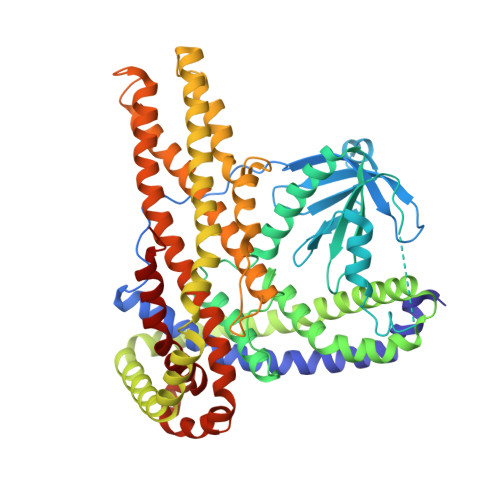
Structural Insight on Functional Regulation of Human MINERVA Protein.
Hahn, H., Lee, D.E., Jang, D.M., Kim, J., Lee, Y., Cheong, H., Han, B.W., Kim, H.S.(2020) Int J Mol Sci 21
- PubMed: 33142954
- DOI: https://doi.org/10.3390/ijms21218186
- Primary Citation of Related Structures:
7CTP - PubMed Abstract:
MINERVA (melanoma invasion by ERK), also known as FAM129B, is a member of the FAM129 protein family, which is only present in vertebrates. MINERVA is involved in key signaling pathways regulating cell survival, proliferation and apoptosis and found upregulated in many types of cancer promoting invasion. However, the exact function of the protein remains elusive. X-ray crystallographic methods were implemented to determine the crystal structure of MINERVA ΔC , lacking C-terminal flexible region. Trypsin digestion was required before crystallization to obtain diffraction-quality crystals. While the N-terminal pleckstrin homology (PH) domain exhibits the typical fold of PH domains, lipid binding assay indicates specific affinity towards phosphatidic acid and inositol 3-phosphate. A helix-rich domain that constitutes the rest of the molecule demonstrates a novel L-shaped fold that encompasses the PH domain. The overall structure of MINERVA ΔC with binding assays and cell-based experiments suggest plasma membrane association of MINERVA and its function seem to be tightly regulated by various motifs within the C-terminal flexible region. Elucidation of MINERVA ΔC structure presents a novel fold for an α-helix bundle domain that would provide a binding platform for interacting partners.
- Research Institute, National Cancer Center, Goyang, Gyeonggi 10408, Korea.
Organizational Affiliation: