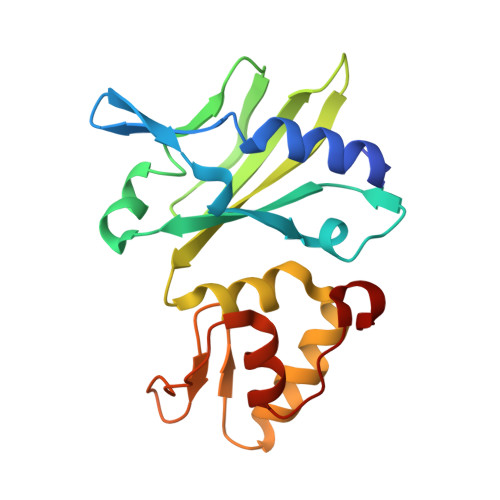
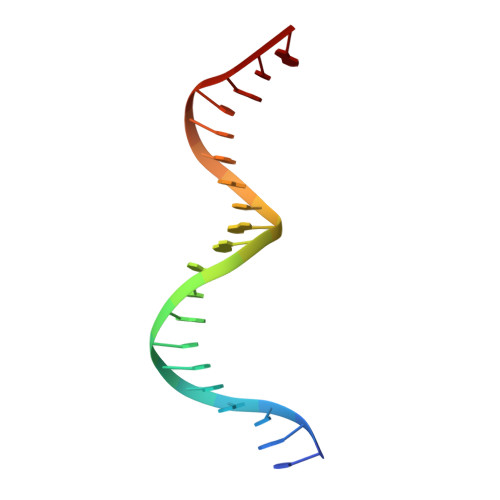
Crystallization and Structural Determination of 8-17 DNAzyme.
Liu, H., Mao, S., Sheng, J., Gan, J.(2022) Methods Mol Biol 2439: 117-130
- PubMed: 35226319
- DOI: https://doi.org/10.1007/978-1-0716-2047-2_9
- Primary Citation of Related Structures:
7CPW - PubMed Abstract:
DNAzymes are a group of DNA molecules that can catalyze various chemical reactions. Owing to their great application potentials, DNAzymes have received significant attention. However, due to their intrinsic difficulties in crystallization and structural determination, only very limited structural information of DNAzymes is available to date. Using co-crystallization with the African Swine Fever Virus Polymerase X (AsfvPolX) protein, we have recently solved a complex structure of the 8-17 DNAzyme, which represents the first structure of the catalytically active RNA-cleaving DNAzyme. In this chapter, we describe the detailed protocols including gene construction, AsfvPolX expression and purification, crystallization, structure determination, and in vitro cleavage assay. While the specific methods described herein were originally designed for the 8-17 DNAzyme, they can also be utilized to solve other DNAzyme structures.
- Shanghai Public Health Clinical Center, State Key Laboratory of Genetic Engineering, Collaborative Innovation Center of Genetics and Development, School of Life Sciences, Fudan University, Shanghai, China.
Organizational Affiliation: