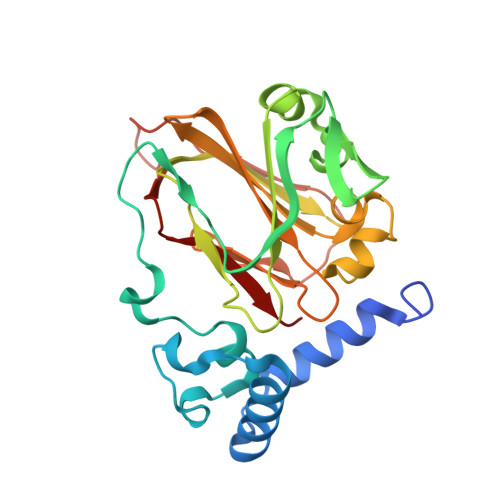
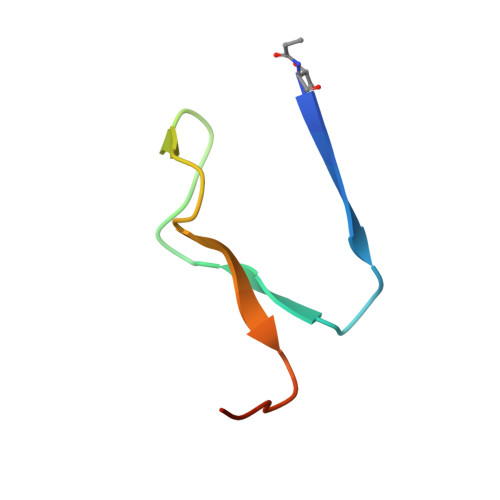
Structural insights into phosphatidylethanolamine formation in bacterial membrane biogenesis.
Cho, G., Lee, E., Kim, J.(2021) Sci Rep 11: 5785-5785
- PubMed: 33707636
- DOI: https://doi.org/10.1038/s41598-021-85195-5
- Primary Citation of Related Structures:
7CNW, 7CNX, 7CNY, 7CNZ - PubMed Abstract:
Phosphatidylethanolamine (PE), a major component of the cellular membrane across all domains of life, is synthesized exclusively by membrane-anchored phosphatidylserine decarboxylase (PSD) in most bacteria. The enzyme undergoes auto-cleavage for activation and utilizes the pyruvoyl moiety to form a Schiff base intermediate with PS to facilitate decarboxylation. However, the structural basis for self-maturation, PS binding, and decarboxylation processes directed by PSD remain unclear. Here, we present X-ray crystal structures of PSD from Escherichia coli, representing an apo form and a PE-bound complex, in which the phospholipid is chemically conjugated to the essential pyruvoyl residue, mimicking the Schiff base intermediate. The high-resolution structures of PE-complexed PSD clearly illustrate extensive hydrophobic interactions with the fatty acyl chains of the phospholipid, providing insights into the broad specificity of the enzyme over a wide range of cellular PS. Furthermore, these structures strongly advocate the unique topology of the enzyme in a lipid bilayer environment, where the enzyme associates with cell membranes in a monotopic fashion via the N-terminal domain composed of three amphipathic helices. Lastly, mutagenesis analyses reveal that E. coli PSD primarily employs D90/D142-H144-S254 to achieve auto-cleavage for the proenzyme maturation, where D90 and D142 act in complementary to each other.
- Department of Chemistry, Gwangju Institute of Science and Technology, Gwangju, 61005, Republic of Korea.
Organizational Affiliation: