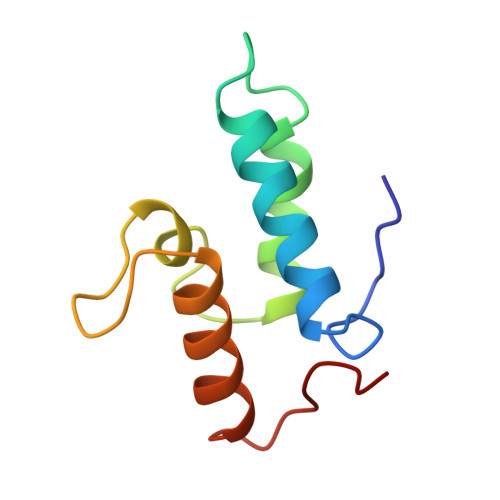
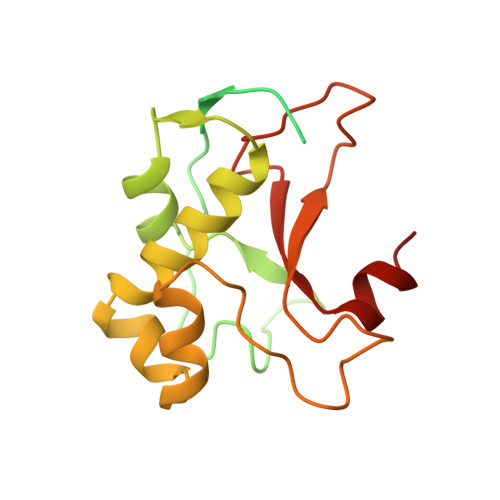
The crystal structure of the DNase domain of colicin E7 in complex with its inhibitor Im7 protein.
Ko, T.P., Liao, C.C., Ku, W.Y., Chak, K.F., Yuan, H.S.(1999) Structure 7: 91-102
- PubMed: 10368275
- DOI: https://doi.org/10.1016/s0969-2126(99)80012-4
- Primary Citation of Related Structures:
7CEI - PubMed Abstract:
Colicin E7 (ColE7) is one of the bacterial toxins classified as a DNase-type E-group colicin. The cytotoxic activity of a colicin in a colicin-producing cell can be counteracted by binding of the colicin to a highly specific immunity protein. This biological event is a good model system for the investigation of protein recognition. The crystal structure of a one-to-one complex between the DNase domain of colicin E7 and its cognate immunity protein Im7 has been determined at 2.3 A resolution. Im7 in the complex is a varied four-helix bundle that is identical to the structure previously determined for uncomplexed Im7. The structure of the DNase domain of ColE7 displays a novel alpha/beta fold and contains a Zn2+ ion bound to three histidine residues and one water molecule in a distorted tetrahedron geometry. Im7 has a V-shaped structure, extending two arms to clamp the DNase domain of ColE7. One arm (alpha1(*)-loop12-alpha2(*); where * represents helices in Im7) is located in the region that displays the greatest sequence variation among members of the immunity proteins in the same subfamily. This arm mainly uses acidic sidechains to interact with the basic sidechains in the DNase domain of ColE7. The other arm (loop 23-alpha3(*)-loop 34) is more conserved and it interacts not only with the sidechain but also with the mainchain atoms of the DNase domain of ColE7. The protein interfaces between the DNase domain of ColE7 and Im7 are charge-complementary and charge interactions contribute significantly to the tight and specific binding between the two proteins. The more variable arm in Im7 dominates the binding specificity of the immunity protein to its cognate colicin. Biological and structural data suggest that the DNase active site for ColE7 is probably near the metal-binding site.
- Institute of Molecular Biology, Academia Sinica, Taipei, Taiwan 11529, Republic of China.
Organizational Affiliation: