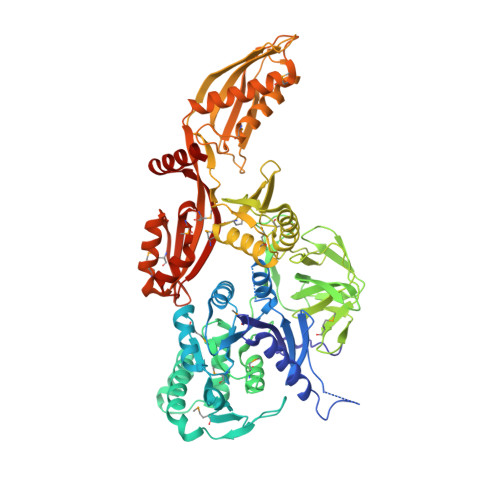
Crystal Structure of Mycobacterium tuberculosis Elongation Factor G1.
Gao, X., Yu, X., Zhu, K., Qin, B., Wang, W., Han, P., Aleksandra Wojdyla, J., Wang, M., Cui, S.(2021) Front Mol Biosci 8: 667638-667638
- PubMed: 34540889
- DOI: https://doi.org/10.3389/fmolb.2021.667638
- Primary Citation of Related Structures:
7CDW - PubMed Abstract:
Mycobacterium tuberculosis ( Mtb ) caused an estimated 10 million cases of tuberculosis and 1.2 million deaths in 2019 globally. The increasing emergence of multidrug-resistant and extensively drug-resistant Mtb is becoming a public health threat worldwide and makes the identification of anti- Mtb drug targets urgent. Elongation factor G (EF-G) is involved in tRNA translocation on ribosomes during protein translation. Therefore, EF-G is a major focus of structural analysis and a valuable drug target of antibiotics. However, the crystal structure of Mtb EF-G1 is not yet available, and this has limited the design of inhibitors. Here, we report the crystal structure of Mtb EF-G1 in complex with GDP. The unique crystal form of the Mtb EF-G1-GDP complex provides an excellent platform for fragment-based screening using a crystallographic approach. Our findings provide a structure-based explanation for GDP recognition, and facilitate the identification of EF-G1 inhibitors with potential interest in the context of drug discovery.
- NHC Key Laboratory of Systems Biology of Pathogens, Institute of Pathogen Biology, And Center for Tuberculosis Research, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China.
Organizational Affiliation: