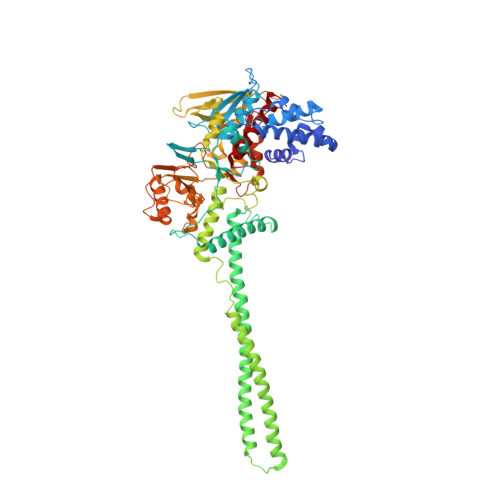
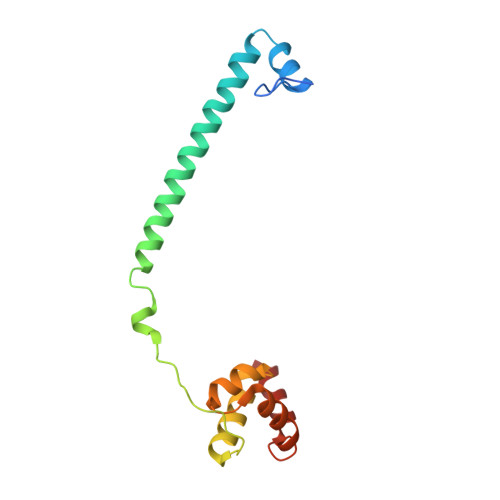
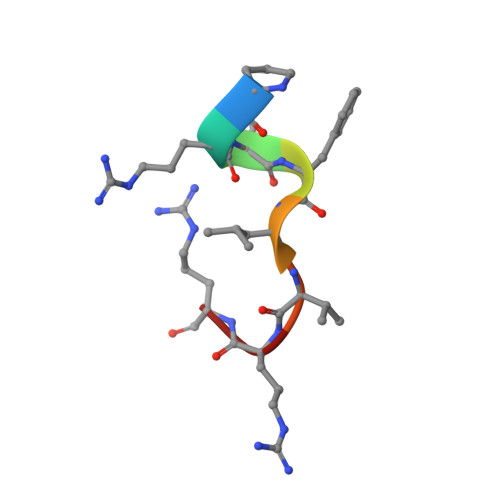
Structure-Based Identification of Potent Lysine-Specific Demethylase 1 Inhibitor Peptides and Temporary Cyclization to Enhance Proteolytic Stability and Cell Growth-Inhibitory Activity.
Kitagawa, H., Kikuchi, M., Sato, S., Watanabe, H., Umezawa, N., Kato, M., Hisamatsu, Y., Umehara, T., Higuchi, T.(2021) J Med Chem 64: 3707-3719
- PubMed: 33754721
- DOI: https://doi.org/10.1021/acs.jmedchem.0c01371
- Primary Citation of Related Structures:
7CDC, 7CDD, 7CDE, 7CDF, 7CDG - PubMed Abstract:
Peptides are attractive drug candidates, but their utility is greatly limited by their inherent susceptibility to proteolytic degradation and their inability to pass through the cell membrane. Here, we employ a strategy of temporary cyclization to develop a cell-active lysine-specific demethylase 1 (LSD1/KDM1A) inhibitor peptide. We first identified a highly potent LSD1-inhibitory linear peptide, with the assistance of X-ray crystal structure data of inhibitor peptide-bound LSD1·CoREST. The peptide was converted to a redox-activatable cyclic peptide incorporating cell-penetrating peptide (CPP), expecting selective activation under intracellular reducing conditions. The cyclic peptide moiety exhibited enhanced stability to protease and was converted to the linear, unmodified LSD1 inhibitor peptide under reducing conditions. The cyclic peptide with CPP inhibited the proliferation of human acute myeloid leukemia cells (HL-60) in the low micromolar concentration range.
- Graduate School of Pharmaceutical Sciences, Nagoya City University, 3-1 Tanabe-dori, Mizuho-ku, Nagoya, Aichi 467-8603, Japan.
Organizational Affiliation: