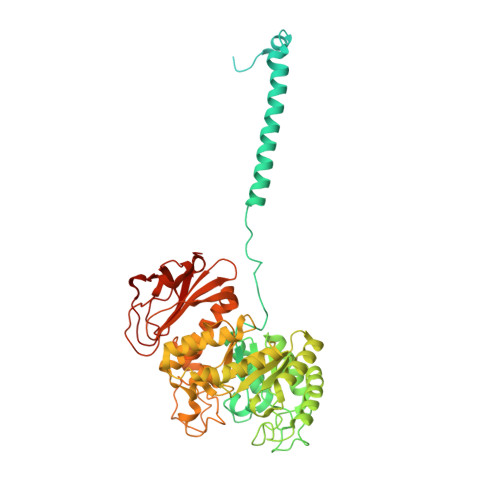
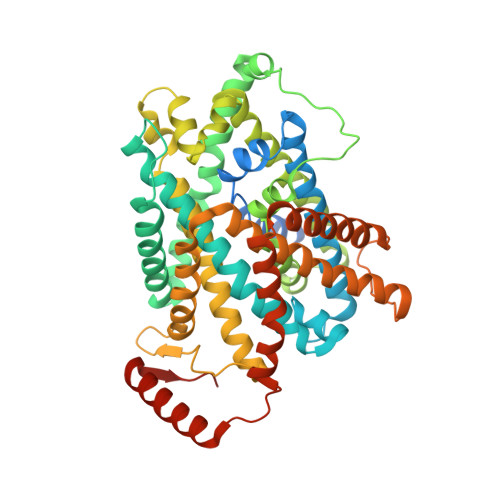
Consensus mutagenesis approach improves the thermal stability of system x c - transporter, xCT, and enables cryo-EM analyses.
Oda, K., Lee, Y., Wiriyasermkul, P., Tanaka, Y., Takemoto, M., Yamashita, K., Nagamori, S., Nishizawa, T., Nureki, O.(2020) Protein Sci 29: 2398-2407
- PubMed: 33016372
- DOI: https://doi.org/10.1002/pro.3966
- Primary Citation of Related Structures:
7CCS - PubMed Abstract:
System x c - is an amino acid antiporter that imports L-cystine into cells and exports intracellular L-glutamate, at a 1:1 ratio. As L-cystine is an essential precursor for glutathione synthesis, system x c - supports tumor cell growth through glutathione-based oxidative stress resistance and is considered as a potential therapeutic target for cancer treatment. System x c - consists of two subunits, the light chain subunit SLC7A11 (xCT) and the heavy chain subunit SLC3A2 (also known as CD98hc or 4F2hc), which are linked by a conserved disulfide bridge. Although the recent structures of another SLC7 member, L-type amino acid transporter 1 (LAT1) in complex with CD98hc, have provided the structural basis toward understanding the amino acid transport mechanism, the detailed molecular mechanism of xCT remains unknown. To revealthe molecular mechanism, we performed single-particle analyses of the xCT-CD98hc complex. As wild-type xCT-CD98hc displayed poor stability and could not be purified to homogeneity, we applied a consensus mutagenesis approach to xCT. The consensus mutated construct exhibited increased stability as compared to the wild-type, and enabled the cryoelectron microscopy (cryo-EM) map to be obtained at 6.2 Å resolution by single-particle analysis. The cryo-EM map revealed sufficient electron density to assign secondary structures. In the xCT structure, the hash and arm domains are well resolved, whereas the bundle domain shows some flexibility. CD98hc is positioned next to the xCT transmembrane domain. This study provides the structural basis of xCT, and our consensus-based strategy could represent a good choice toward solving unstable protein structures.
- Department of Biological Sciences, Graduate School of Science, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo, Japan.
Organizational Affiliation: